Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for their Preventive Effects against Salmonella Typhimurium
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeasts, Bacteria Strains, and Growth Conditions
2.2. Human Cell Cultures
2.3. Determination of Acid pH and Bile Tolerance of Yeasts
2.4. In Vitro Yeasts Adhesion Assay
2.5. Growth Inhibition of Salmonella Typhimurium by Yeasts
2.6. Caco-2 TransEpithelial Electrical Resistance (TEER) Assay
2.7. C. elegans Maintenance and Synchronization
2.8. C. elegans Longevity Assay
2.9. C. elegans Survival Assay
2.10. Effects of D. hansenii on S. Typhimurium Infection in the C. elegans Survival Assay
2.11. Statistical Analysis
3. Results and discussion
3.1. Acidic pH and Bile Tolerance
3.2. In Vitro Adhesion Assay
3.3. Growth Inhibition of Salmonella Typhimurium by Yeasts
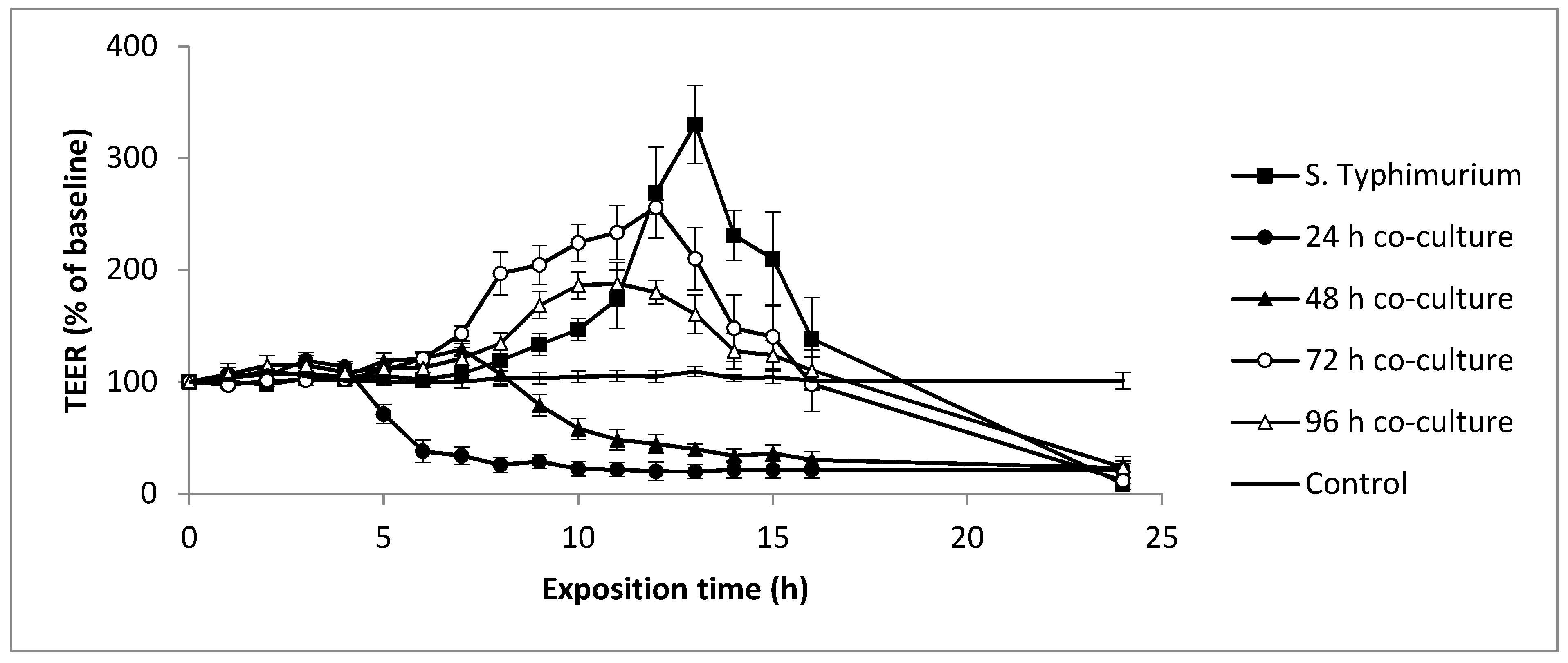
3.4. Evolution of Yeasts and Salmonella Typhimurium-Induced Epithelial Caco-2 Cell Barrier Function
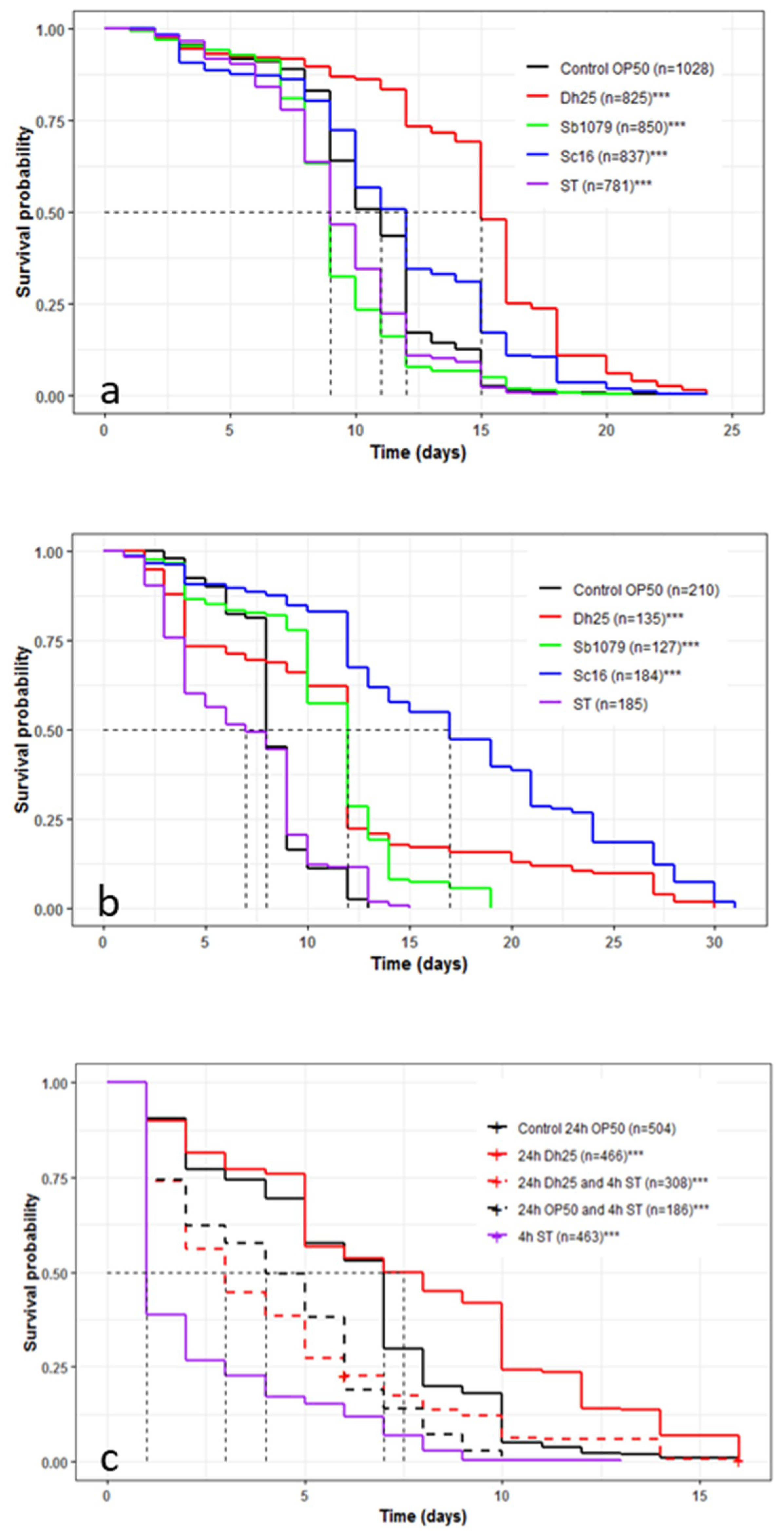
3.5. Influence of Yeasts and Salmonella Typhimurium on the Longevity and Survival of Caenorhabditis elegans and the Preventive Effects of Debaryomyces hansenii 25
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| TEER | TransEpithelial Electrical Resistance |
References
- Huffnagle, G.B.; Noverr, M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013, 21, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Lukeš, J.; Stensvold, C.R.; Jirků-Pomajbíková, K.; Wegener Parfrey, L. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 2015, 11, e1005039. [Google Scholar]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The human microbiota in health and disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- FAO; WHO. Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria. Food Nutr. Pap. 2001, 85, 71. [Google Scholar]
- Didari, T.; Solki, S.; Mozaffari, S.; Nikfar, S.; Abdollahi, M. A systematic review of the safety of probiotics. Expert Opin. Drug Saf. 2014, 13, 227–239. [Google Scholar] [CrossRef]
- Dinleyici, E.C.; Eren, M.; Ozen, M.; Yargic, Z.A.; Vandenplas, Y. Effectiveness and safety of Saccharomyces boulardii for acute infectious diarrhea. Expert Opin. Biol. Ther. 2012, 12, 395–410. [Google Scholar] [CrossRef]
- McFarland, L.V. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World J. Gastroenterol. 2010, 16, 2202–2222. [Google Scholar] [CrossRef]
- Zanello, G.; Meurens, F.; Berri, M.; Chevaleyre, C.; Melo, S.; Auclair, E.; Salmon, H. Saccharomyces cerevisiae decreases inflammatory responses induced by F4+ enterotoxigenic Escherichia coli in porcine intestinal epithelial cells. Vet. Immunol. Immunopathol. 2011, 141, 133–138. [Google Scholar] [CrossRef]
- Živković, M.; Čadež, N.; Uroić, K.; Miljković, M.; Tolinački, M.; Doušova, P.; Kos, B.; Šušković, J.; Raspor, P.; Topisirović, L.; et al. Evaluation of probiotic potential of yeasts isolated from traditional cheeses manufactured in Serbia and Croatia. J. Intercult. Ethnopharmacol. 2015, 4, 12–18. [Google Scholar] [CrossRef]
- Ochangco, H.S.; Gamero, A.; Smith, I.M.; Christensen, J.E.; Jespersen, L.; Arneborg, N. In vitro investigation of Debaryomyces hansenii strains for potential probiotic properties. World J. Microbiol. Biotechnol. 2016, 32, 141. [Google Scholar] [CrossRef]
- Kunyeit, L.; Kurrey, N.K.; Anu-Appaiah, K.A.; Rao, R.P. Probiotic yeasts inhibit virulence of non-albicans Candida Species. mBIO 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Retureau, E.; Callon, C.; Didienne, R.; Montel, M.-C. Is microbial diversity an asset for inhibiting Listeria monocytogenes in raw milk cheeses? Dairy Sci. Technol. 2010, 90, 375–398. [Google Scholar] [CrossRef]
- Callon, C.; Retureau, E.; Didienne, R.; Montel, M.-C. Microbial biodiversity in cheese consortia and comparative Listeria growth on surfaces of uncooked pressed cheeses. Int. J. Food Microbiol. 2014, 174, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Andreoletti, O.; Budka, H.; Buncic, S.; Colin, P.; Collins, J.D.; De Koeijer. The maintenance of the list of QPS microorganisms intentionally added to food or feed - Scientific Opinion of the Panel on Biological Hazards. EFSA J. 2008, 923, 1–48. [Google Scholar]
- Martins, F.S.; Vieira, A.T.; Elian, S.D.A.; Arantes, R.M.E.; Tiago, F.C.P.; Sousa, L.P.; Araújo, H.R.; Pimenta, P.F.; Bonjardim, C.A.; Nicoli, J.R.; et al. Inhibition of tissue inflammation and bacterial translocation as one of the protective mechanisms of Saccharomyces boulardii against Salmonella infection in mice. Microbes Infect. 2013, 15, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, V.H.; Ishikawa, K.H.; Ando-Suguimoto, E.S.; Bueno-Silva, B.; Nakamae, A.E.M.; Mayer, M.P.A. Probiotic bacteria alter pattern-recognition receptor expression and cytokine profile in a human macrophage model challenged with Candida albicans and lipopolysaccharide. Front. Microbiol. 2017, 8, 2280. [Google Scholar] [CrossRef]
- Leão, M.V.P.; Tavares, T.A.A.; Gonçalves, E.; Silva, C.R.; Dos Santos, S.S.F.; Junqueira, J.C.; de Oliveira, L.D.; Jorge, A.O.C. Lactobacillus rhamnosus intake can prevent the development of candidiasis. Clin. Oral Investig. 2018, 22, 2511–2518. [Google Scholar] [CrossRef]
- Tallon, R.; Arias, S.; Bressollier, P.; Urdaci, M.C. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 2007, 102, 442–451. [Google Scholar] [CrossRef]
- Zielińska, D.; Długosz, E.; Zawistowska-Deniziak, A. Functional properties of food origin Lactobacillus in the gastrointestinal ecosystem—In vitro study. Probiotics Antimicrob. Proteins 2019, 11, 820–829. [Google Scholar] [CrossRef]
- Lacroix, C.; de Wouters, T.; Chassard, C. Integrated multi-scale strategies to investigate nutritional compounds and their effect on the gut microbiota. Curr. Opin. Biotechnol. 2015, 32, 149–155. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J. The C. elegans Intestine; WormBook, Ed.; The C. elegans Research Community, WormBook, 2007; Available online: http://www.wormbook.org/chapters/www_intestine/intestine.html (accessed on 19 May 2020).
- Park, M.R.; Yun, H.S.; Son, S.J.; Oh, S.; Kim, Y. Short communication: Development of a direct in vivo screening model to identify potential probiotic bacteria using Caenorhabditis elegans. J. Dairy Sci. 2014, 97, 6828–6834. [Google Scholar] [CrossRef]
- Lee, J.; Choe, J.; Kim, J.; Oh, S.; Park, S.; Kim, S.; Kim, Y. Heat-killed Lactobacillus spp. cells enhance survivals of Caenorhabditis elegans against Salmonella and Yersinia infections. Lett. Appl. Microbiol. 2015, 61, 523–530. [Google Scholar] [CrossRef]
- Mylonakis, E.; Ausubel, F.M.; Perfect, J.R.; Heitman, J.; Calderwood, S.B. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 15675–15680. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Yun, M.; Politz, S.M.; Rao, R.P. A Pathogenesis Assay Using Saccharomyces cerevisiae and Caenorhabditis elegans reveals novel roles for yeast AP-1, Yap1, and host dual oxidase BLI-3 in fungal pathogenesis. Eukaryot. Cell 2009, 8, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Ausubel, F.M.; Mylonakis, E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011, 7, e1002074. [Google Scholar] [CrossRef] [PubMed]
- Madende, M.; Albertyn, J.; Sebolai, O.; Pohl, C.H. Caenorhabditis elegans as a model animal for investigating fungal pathogenesis. Med. Microbiol. Immunol. 2020, 209, 1–13. [Google Scholar] [CrossRef]
- Romanin, D.E.; Llopis, S.; Genovés, S.; Martorell, P.; Ramón, V.d.; Garrote, G.l.; Rumbo, M. Probiotic yeast Kluyveromyces marxianus CIDCA 8154 shows anti-inflammatory and anti-oxidative stress properties in in vivo models. Benef. Microbes 2015, 7, 83–93. [Google Scholar] [CrossRef]
- Oliveira, C.J.B.; Hisrich, E.R.; Moura, J.F.P.; Givisiez, P.E.N.; Costa, R.G.; Gebreyes, W.A. On farm risk factors associated with goat milk quality in Northeast Brazil. Small Ruminant Res. 2011, 98, 64–69. [Google Scholar] [CrossRef]
- Pointon, A.; Kiermeier, A.; Fegan, N. Review of the impact of pre-slaughter feed curfews of cattle, sheep and goats on food safety and carcase hygiene in Australia. Food Control. 2012, 26, 313–321. [Google Scholar] [CrossRef]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta – Biomembr. 2009, 1788, 832–841. [Google Scholar] [CrossRef]
- Gagnon, M.; Berner, A.Z.; Chervet, N.; Chassard, C.; Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Meth. 2013, 94, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Zweibaum, A.; Laburthe, M.; Grasset, E.; Louvard, D. Use of cultured cell lines in studies of intestinal cell differentiation and function. Compr. Physiol. 2011, 223–255. [Google Scholar]
- Pinto, M.; Robine-Leon, S.; Appay, M.D.; Kedinger, M.; Triadou, N.; Dussaulx, E.; Lacroix, B.; Simon-Assmann, P.; Haffen, K.; Fogh, J.; et al. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell. 1983, 47, 323–330. [Google Scholar]
- Nivoliez, A.; Camares, O.; Paquet-Gachinat, M.; Bornes, S.; Forestier, C.; Veisseire, P. Influence of manufacturing processes on in vitro properties of the probiotic strain Lactobacillus rhamnosus Lcr35®. J. Biotechnol. 2012, 160, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Poupet, C.; Saraoui, T.; Veisseire, P.; Bonnet, M.; Dausset, C.; Gachinat, M.; Camarès, O.; Chassard, C.; Nivoliez, A.; Bornes, S. Lactobacillus rhamnosus Lcr35 as an effective treatment for preventing Candida albicans infection in the invertebrate model Caenorhabditis elegans: First mechanistic insights. PLoS ONE 2019, 14, e0216184. [Google Scholar]
- de Barros, P.; Scorzoni, L.; Ribeiro, F.; Fugisaki, L.; Fuchs, B.; Mylonakis, E.; Jorge, A.; Junqueira, J.; Rossoni, R. Lactobacillus paracasei 28.4 reduces in vitro hyphae formation of Candida albicans and prevents the filamentation in an experimental model of Caenorhabditis elegans. Microb. Pathog. 2018, 117, 80–87. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environnement for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org/ (accessed on 19 May 2020).
- Therneau, T.M. A package for survival analysis in R. 2015. Available online: https://github.com/therneau/survival (accessed on 19 May 2020).
- Kassambara, A.; Kosinski, M. Survminer: Drawing Survival Curves Using “ggplot2”. 2017. Available online: http://www.sthda.com/english/rpkgs/survminer/ (accessed on 19 May 2020).
- Edwards-Ingram, L.; Gitsham, P.; Burton, N.; Warhurst, G.; Clarke, I.; Hoyle, D.; Oliver, S.G.; Stateva, L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007, 73, 2458–2467. [Google Scholar] [CrossRef]
- Villar-Garcia, J.; Güerri-Fernandez, R.; Moya, A.; Gonzalez, A.; Hernandez, J.J.; Lerma, E.; Guelar, A.; Sorli, L.; Horcajada, J.P.; Artacho, A.; et al. Impact of probiotic Saccharomyces boulardii on the gut microbiome composition in HIV-treated patients: A double-blind, randomised, placebo-controlled trial. PLoS ONE 2017, 12, e0173802. [Google Scholar] [CrossRef]
- Arévalo-Villena, M.; Fernandez-Pacheco, P.; Castillo, N.; Bevilacqua, A.; Briones Pérez, A. Probiotic capability in yeasts: Set-up of a screening method. LWT 2018, 89, 657–665. [Google Scholar] [CrossRef]
- Asmat, S.; Shaukat, F.; Asmat, R.; Faiq Siddique Gul Bakhat, H.; Asmat, T.M. Clinical efficacy comparison of Saccharomyces boulardii and lactic acid as probiotics in acute pediatric diarrhea. J. Coll. Physicians Surg. Pak. 2018, 28, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Hyronimus, B.; Le Marrec, C.; Sassi, A.H.; Deschamps, A. Acid and bile tolerance of spore-forming lactic acid bacteria. Int. J. Food Microbiol. 2000, 61, 193–197. [Google Scholar] [CrossRef]
- Pedersen, L.L.; Owusu-Kwarteng, J.; Thorsen, L.; Jespersen, L. Biodiversity and probiotic potential of yeasts isolated from Fura, a West African spontaneously fermented cereal. Int. J. Food Microbiol. 2012, 159, 144–151. [Google Scholar] [CrossRef]
- Trotta, F.; Caldini, G.; Dominici, L.; Federici, E.; Tofalo, R.; Schirone, M.; Corsetti, A.; Suzzi, G.; Cenci, G. Food borne yeasts as DNA-bioprotective agents against model genotoxins. Int. J. Food Microbiol. 2012, 153, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-S.; Ma, Y.; Maubois, J.-L.; He, S.-H.; Chen, L.-J.; Li, H.-M. Screening for the potential probiotic yeast strains from raw milk to assimilate cholesterol. Dairy Sci. Technol. 2010, 90, 537–548. [Google Scholar] [CrossRef]
- Lange, K. Fundamental role of microvilli in the main functions of differentiated cells: Outline of a universal regulating and signaling system at the cell periphery. J. Cell Physiol. 2011, 226, 896–927. [Google Scholar] [CrossRef]
- Espey, M.G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 2013, 55, 130–140. [Google Scholar] [CrossRef]
- Lima, M.D.S.F.; de Souza, K.M.S.; de Albuquerque, W.W.C.; Teixeira, J.A.C.; Cavalcanti, M.T.H.; Porto, A.L.F. Saccharomyces cerevisiae from Brazilian kefir-fermented milk: An in-vitro evaluation of probiotic properties. Microb. Pathog. 2017, 110, 670–677. [Google Scholar] [CrossRef]
- Roussel, C.; Sivignon, A.; de Vallée, A.; Garrait, G.; Denis, S.; Tsilia, V.; Ballet, N.; Vandekerckove, P.; Van de Wiele, T.; Barnich, N.; et al. Anti-infectious properties of the probiotic Saccharomyces cerevisiae CNCM I-3856 on enterotoxigenic E. coli (ETEC) strain H10407. Appl. Microbiol. Biotechnol. 2018, 102, 6175–6189. [Google Scholar] [CrossRef]
- Binetti, A.; Carrasco, M.; Reinheimer, J.; Suárez, V. Yeasts from autochthonal cheese starters: Technological and functional properties. J. Appl. Microbiol. 2013, 115, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Perricone, M.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Technological characterization and probiotic traits of yeasts isolated from Altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 2014, 38, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Klingberg, T.D.; Pedersen, M.H.; Cencic, A.; Budde, B.B. Application of Measurements of Transepithelial Electrical Resistance of Intestinal Epithelial Cell Monolayers to Evaluate Probiotic Activity. Appl. Environ. Microbiol. 2005, 71, 7528–7530. [Google Scholar] [CrossRef] [PubMed]
- Sambuy, Y.; Angelis, I.D.; Ranaldi, G.; Scarino, M.L.; Stammati, A.; Zucco, F. The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell. Biol. Toxicol. 2005, 21, 1–26. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Smith, I.M.; Baker, A.; Arneborg, N.; Jespersen, L. Non-Saccharomyces yeasts protect against epithelial cell barrier disruption induced by Salmonella enterica subsp. enterica serovar Typhimurium. Lett. Appl. Microbiol. 2015, 61, 491–497. [Google Scholar] [CrossRef]
- Knodler, L.A.; Vallance, B.A.; Celli, J.; Winfree, S.; Hansen, B.; Montero, M.; Steele-Mortimer, O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. USA 2010, 107, 17733–17738. [Google Scholar] [CrossRef] [PubMed]
- Oza, J.P.; Yeh, J.B.; Reich, N.O. DNA methylation modulates Salmonella enterica serovar Typhimurium virulence in Caenorhabditis elegans. FEMS Microbiol. Lett. 2005, 245, 53–59. [Google Scholar] [CrossRef][Green Version]
- Sahu, S.N.; Anriany, Y.; Grim, C.J.; Kim, S.; Chang, Z.; Joseph, S.W.; Cinar, H.N. (2013) Identification of virulence properties in Salmonella Typhimurium DT104 using Caenorhabditis elegans. PLoS ONE 2005, 8, e76673. [Google Scholar]
- Zhou, M.; Yu, H.; Yin, X.; Sabour, P.M.; Chen, W.; Gong, J. Lactobacillus zeae protects Caenorhabditis elegans from Enterotoxigenic Escherichia coli-caused death by inhibiting enterotoxin gene expression of the pathogen. PLoS ONE 2014, 9, e89004. [Google Scholar] [CrossRef]
- Tullet, J.M.A. DAF-16 target identification in C. elegans: Past, present and future. Biogerontology 2015, 16, 221–234. [Google Scholar] [CrossRef] [PubMed]

| a. Survival rates in the presence of acidic pH (%) | ||||
| Strains | 45 min | 90 min | ||
| S. boulardii 1079 | 93.6 ± 8.3 | 91.1 ± 9.0 | ||
| D. hansenii 25 | 96.6 ± 3.4 | 85.5 ± 3.0 | ||
| S. cerevisiae 16 | 101.3 ± 3.4 | 54.2 ± 2.7 | ||
| b. Survival rates in the presence of bile salts (%) | ||||
| Strains | 1 h | 2 h | 3 h | 4 h |
| S. boulardii 1079 | 85.3 ± 4.5 | 70.3 ± 4.4 | 63.1 ± 2.7 | 73.5 ± 4.9 |
| D. hansenii 25 | 94.1 ± 7.4 | 91.3 ± 7.7 | 72.8 ± 13.5 | 68.9 ± 16.1 |
| S. cerevisiae 16 | 89.2 ± 2.9 | 77.5 ± 15.1 | 56.0 ± 9.4 | 39.2 ± 6.7 |
| a. Multiplicity of infection (MOI) | ||||
| HT29-MTX | MOI 100 | MOI 10 | MOI 1 | MOI 0.1 |
| S. boulardii 1079 | 0.51 ± 0.046% a,b | 0.39 ± 0.123% a,b | 0.46 ± 0.189% a,b | 1.15 ± 0.270% a,b |
| D. hansenii 25 | 0.18 ± 0.005% b | 0.29 ± 0.005% b | 0.44 ± 0.007% b | 1.56 ± 0.010% b |
| S. cerevisiae 16 | 0.91 ± 0.015% a | 0.90 ± 0.013% a | 1.24 ± 0.020% a | 2.61 ± 0.024% a |
| b. Multiplicity of infection (MOI) | ||||
| Caco-2 | MOI 100 | MOI 10 | MOI 1 | MOI 0.1 |
| S. boulardii 1079 | 0.59 ± 0.006% a | 1.87 ± 0.011% a | 1.77 ± 0.013% a,b | 3.60 ± 0.028% a |
| D. hansenii 25 | 2.27 ± 0.011% b | 2.50 ± 0.060% a | 4.10 ± 0.325% b | 4.12 ± 1.701% a |
| S. cerevisiae 16 | 0.81 ± 0.009% a | 0.86 ± 0.007% b | 2.56 ± 0.032% a | 7.26 ± 0.088% a |
| Time (h) | 0 | 24 | 48 |
|---|---|---|---|
| ST | 6.47 ± 0.48 a | 8.88 ± 0.30 a | 8.71 ± 0.20 a |
| ST + Sb1079 | 6.63 ± 0.38 a | 8.39 ± 0.50 a,b | 6.46 ± 0.92 b |
| ST + Dh25 | 6.58 ± 0.46 a | 8.34 ± 0.62 b | 6.90 ± 0.97 b |
| ST + Sc16 | 6.60 ± 0.43 a | 8.21 ± 0.43 b | 6.91 ± 0.97 b |
| Co-Culture Duration | ST (log CFU/mL) | Dh25 (log CFU/mL) |
|---|---|---|
| 0 h | 6.40 ± 0.10 a | 5.93 ± 0.21 a |
| 24 h | 8.08 ± 0.58 b | 6.60 ± 0.35 a |
| 48 h | 6.90 ± 0.13 a | 5.90 ± 0.56 a |
| 72 h | 6.85 ± 0.30 a | 4.87 ± 0.81 b |
| 96 h | 6.79 ± 0.09 a | 4.15 ± 0.85 c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veisseire, P.; Bonnet, M.; Saraoui, T.; Poupet, C.; Camarès, O.; Gachinat, M.; Callon, C.; Febvre, G.; Chassard, C.; Bornes, S. Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for their Preventive Effects against Salmonella Typhimurium. Microorganisms 2020, 8, 922. https://doi.org/10.3390/microorganisms8060922
Veisseire P, Bonnet M, Saraoui T, Poupet C, Camarès O, Gachinat M, Callon C, Febvre G, Chassard C, Bornes S. Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for their Preventive Effects against Salmonella Typhimurium. Microorganisms. 2020; 8(6):922. https://doi.org/10.3390/microorganisms8060922
Chicago/Turabian StyleVeisseire, Philippe, Muriel Bonnet, Taous Saraoui, Cyril Poupet, Olivier Camarès, Marylise Gachinat, Cécile Callon, Guy Febvre, Christophe Chassard, and Stéphanie Bornes. 2020. "Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for their Preventive Effects against Salmonella Typhimurium" Microorganisms 8, no. 6: 922. https://doi.org/10.3390/microorganisms8060922
APA StyleVeisseire, P., Bonnet, M., Saraoui, T., Poupet, C., Camarès, O., Gachinat, M., Callon, C., Febvre, G., Chassard, C., & Bornes, S. (2020). Investigation into In Vitro and In Vivo Caenorhabditis elegans Models to Select Cheese Yeasts as Probiotic Candidates for their Preventive Effects against Salmonella Typhimurium. Microorganisms, 8(6), 922. https://doi.org/10.3390/microorganisms8060922





