A Rapid, Sensitive, Low-Cost Assay for Detecting Hydrogenotrophic Methanogens in Anaerobic Digesters Using Loop-Mediated Isothermal Amplification
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of LAMP Assay Biomarkers
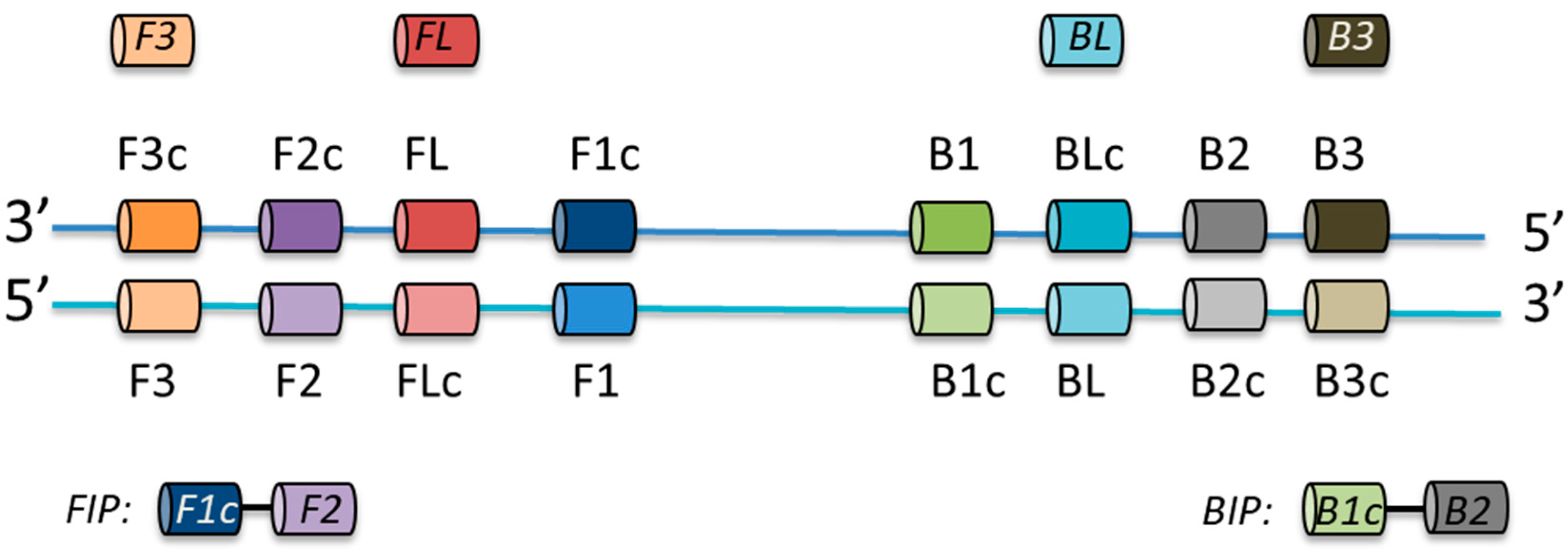
2.2. LAMP Primer Design
2.3. Template DNA
2.4. PCR with Outer F3/B3 Primers
2.5. LAMP Assay
2.6. LAMP Sensitivity
3. Results
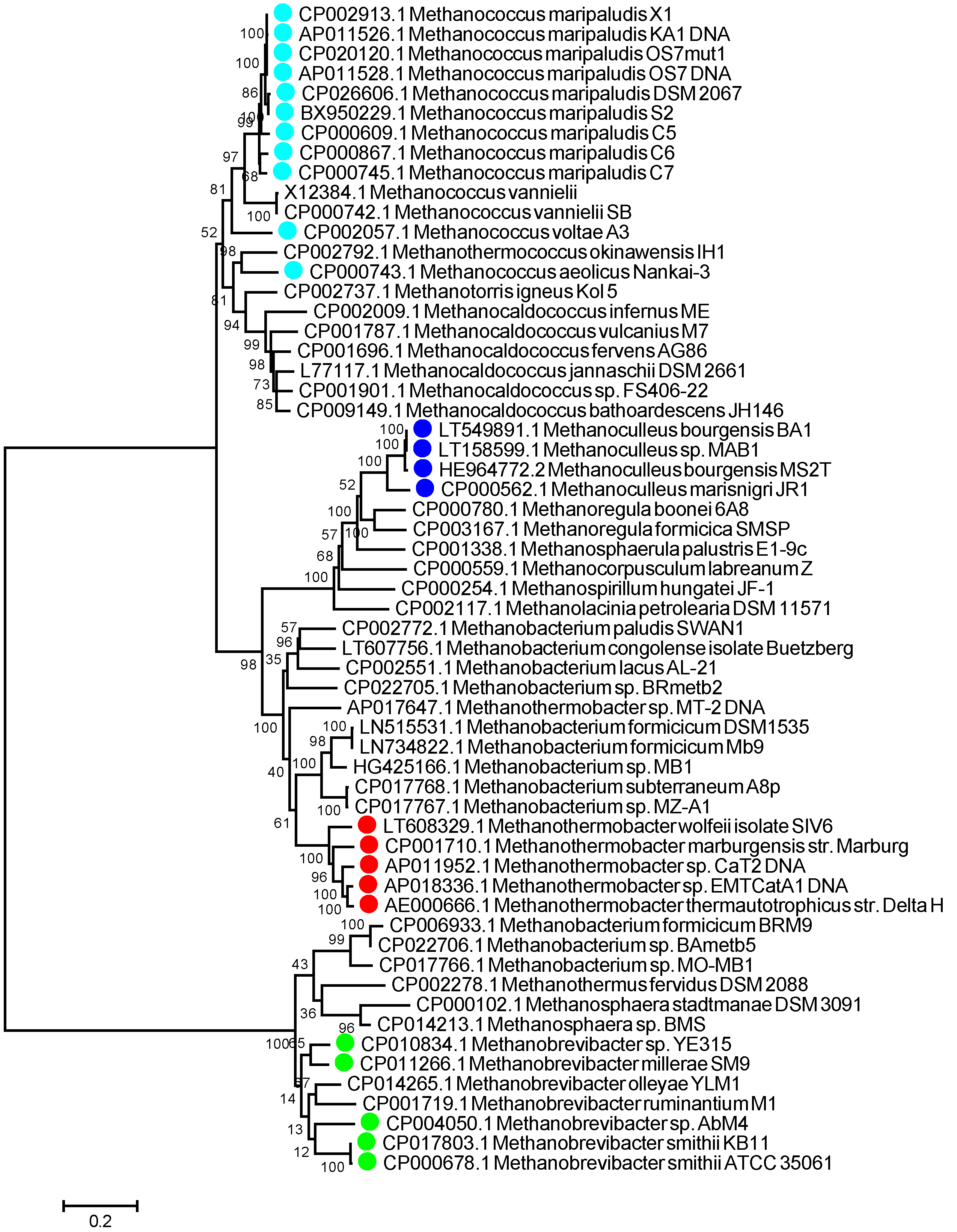
3.1. aEF-2 Is a Potential Biomarker for Hydrogenotrophic Methanogen Detection in LAMP Assays
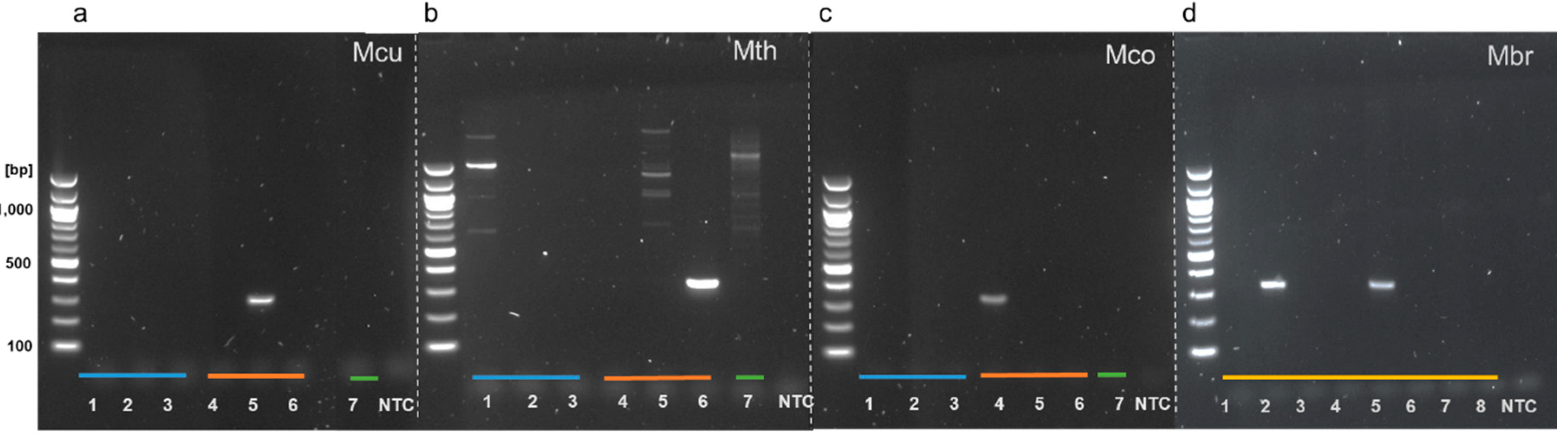
3.2. Genus-Specific PCR Products Are Generated with LAMP Outer Primers
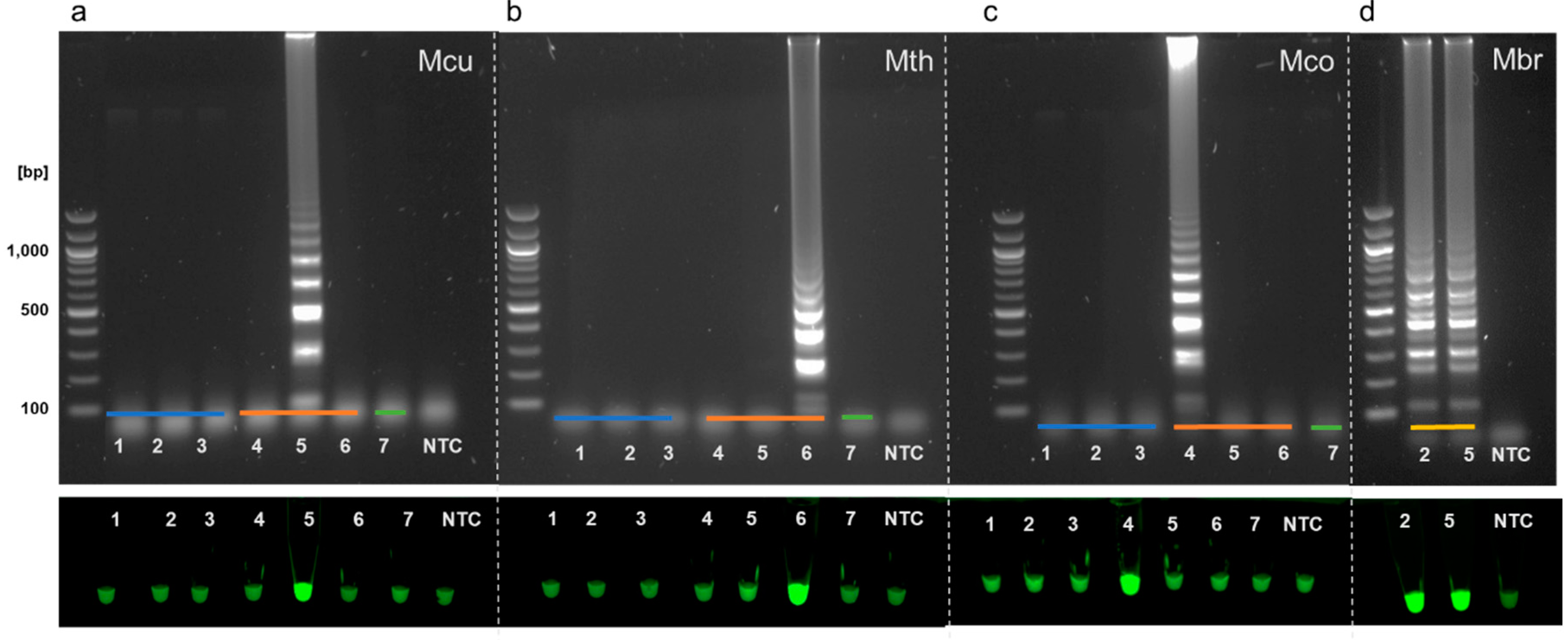
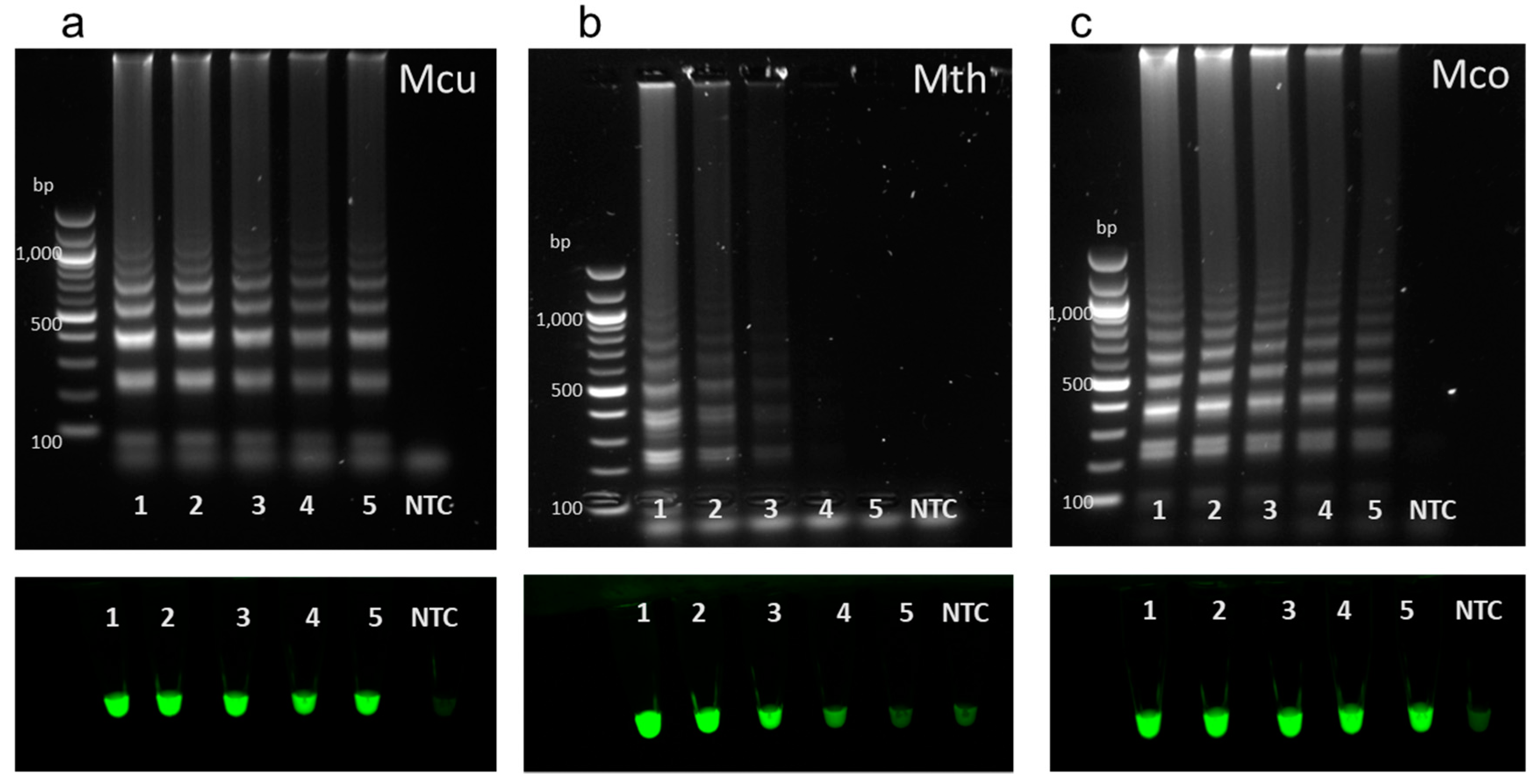
3.3. Specificity of Full LAMP Assays
3.4. A Set of LAMP Assays to Discriminate between Hydrogenotrophic Methanogenic Genera
3.5. LAMP Sensitivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kougias, P.G.; Angelidaki, I. Biogas and Its Opportunities—A Review. Front. Environ. Sci. Eng. 2018, 12, 14. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.-K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic Archaea: Ecologically Relevant Differences in Energy Conservation. Nat. Rev. Genet. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Galand, P.E.; Fritze, H.; Conrad, R.; Yrjälä, K. Pathways for Methanogenesis and Diversity of Methanogenic Archaea in Three Boreal Peatland Ecosystems. Appl. Environ. Microbiol. 2005, 71, 2195–2198. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Moestedt, J.; Schnürer, A. Biogas Production through Syntrophic Acetate Oxidation and Deliberate Operating Strategies for Improved Digester Performance. Appl. Energy 2016, 179, 124–135. [Google Scholar] [CrossRef]
- Tao, B.; Alessi, A.M.; Zhang, Y.; Chong, J.P.; Heaven, S.; Banks, C.J. Simultaneous Biomethanisation of Endogenous and Imported CO2 in Organically Loaded Anaerobic Digesters. Appl. Energy 2019, 247, 670–681. [Google Scholar] [CrossRef]
- Angelidaki, I.; Treu, L.; Tsapekos, P.; Luo, G.; Campanaro, S.; Wenzel, H.; Kougias, P.G. Biogas Upgrading and Utilization: Current Status and Perspectives. Biotechnol. Adv. 2018, 36, 452–466. [Google Scholar] [CrossRef]
- Bassani, I.; Kougias, P.G.; Treu, L.; Angelidaki, I. Biogas Upgrading via Hydrogenotrophic Methanogenesis in Two-Stage Continuous Stirred Tank Reactors at Mesophilic and Thermophilic Conditions. Environ. Sci. Technol. 2015, 49, 12585–12593. [Google Scholar] [CrossRef]
- Kougias, P.G.; Treu, L.; Benavente, D.P.; Boe, K.; Campanaro, S.; Angelidaki, I. Ex-Situ Biogas Upgrading and Enhancement in Different Reactor Systems. Bioresour. Technol. 2017, 225, 429–437. [Google Scholar] [CrossRef]
- Müller, B.; Sun, L.; Westerholm, M.; Schnürer, A. Bacterial Community Composition and Fhs Profiles of Low- and High-Ammonia Biogas Digesters Reveal Novel Syntrophic Acetate-Oxidising Bacteria. Biotechnol. Biofuels 2016, 9, 48. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of Previously Uncultured Members of the Human Gut Microbiota by Culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.H.; Masubuchi, T.; Yonekawa, K.; Amino, W.N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.-P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.; Son, R. Loop-Mediated Isothermal Amplification (LAMP): A Versatile Technique for Detection of Micro-Organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Kimura, Y.; de Hoon, M.J.L.; Aoki, S.; Ishizu, Y.; Kawai, Y.; Kogo, Y.; Daub, C.O.; Lezhava, A.; Arner, E.; Hayashizaki, Y. Optimization of Turn-Back Primers in Isothermal Amplification. Nucleic Acids Res. 2011, 39, e59. [Google Scholar] [CrossRef]
- Romesser, J.A.; Wolfe, R.S.; Mayer, F.; Spiess, E.; Walther-Mauruschat, A. Methanogenium, a New Genus of Marine Methanogenic Bacteria, and Characterization of Methanogenium cariaci Sp. Nov. and Methanogenium marisnigri Sp. Nov. Arch. Microbiol. 1979, 121, 147–153. [Google Scholar] [CrossRef]
- Haydock, A.K.; Porat, I.; Whitman, B.; Leigh, J.A. Continuous Culture of Methanococcus Maripaludis under Defined Nutrient Conditions. FEMS Microbiol. Lett. 2004, 238, 85–91. [Google Scholar] [CrossRef][Green Version]
- Nölling, J.; Frijlink, M.; De Vos, W.M. Isolation and Characterization of Plasmids from Different Strains of Methanobacterium Thermoformicicum. Microbiology 1991, 137, 1981–1986. [Google Scholar] [CrossRef][Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Karthik, K.; Rathore, R.; Thomas, P.; Arun, T.; Viswas, K.; Dhama, K.; Agarwal, R. New Closed Tube Loop Mediated Isothermal Amplification Assay for Prevention of Product Cross-Contamination. MethodsX 2014, 1, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Gupta, R.S. Phylogenomic Analysis of Proteins That Are Distinctive of Archaea and Its Main Subgroups and the Origin of Methanogenesis. BMC Genom. 2007, 8, 86. [Google Scholar] [CrossRef]
- Friedrich, M.W. Methyl-Coenzyme M Reductase Genes: Unique Functional Markers for Methanogenic and Anaerobic Methane-Oxidizing Archaea. Methods Enzymol. 2005, 397, 428–442. [Google Scholar] [CrossRef]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical Background and Biotechnological Applications. AMB Express 2018, 8, 1. [Google Scholar] [CrossRef]
- Iwabe, N.; Kuma, K.; Hasegawa, M.; Osawa, S.; Miyata, T. Evolutionary Relationship of Archaebacteria, Eubacteria, and Eukaryotes Inferred from Phylogenetic Trees of Duplicated Genes. Proc. Natl. Acad. Sci. USA 1989, 86, 9355–9359. [Google Scholar] [CrossRef]
- Elkins, J.G.; Podar, M.L.; Graham, D.E.; Makarova, K.S.; Wolf, Y.; Randau, L.; Hedlund, B.P.; Brochier-Armanet, C.; Kunin, V.; Anderson, I.; et al. A Korarchaeal Genome Reveals Insights into the Evolution of the Archaea. Proc. Natl. Acad. Sci. USA 2008, 105, 8102–8107. [Google Scholar] [CrossRef]
- Williams, T.A.; Foster, P.G.; Nye, T.M.W.; Cox, C.J.; Embley, T.M. A Congruent Phylogenomic Signal Places Eukaryotes within the Archaea. Proc. R. Soc. B Boil. Sci. 2012, 279, 4870–4879. [Google Scholar] [CrossRef]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The Two-Domain Tree of Life Is Linked to a New Root for the Archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef]
- Narrowe, A.B.; Spang, A.; Stairs, C.W.; Caceres, E.F.; Baker, B.J.; Miller, C.S.; Ettema, T.J.G. Complex Evolutionary History of Translation Elongation Factor 2 and Diphthamide Biosynthesis in Archaea and Parabasalids. Genome Biol. Evol. 2018, 10, 2380–2393. [Google Scholar] [CrossRef]
- Kirkegaard, R.H.; McIlroy, S.J.; Kristensen, J.M.; Nierychlo, M.; Karst, S.M.; Dueholm, M.S.; Albertsen, M.; Nielsen, P.H. The Impact of Immigration on Microbial Community Composition in Full-Scale Anaerobic Digesters. Sci. Rep. 2017, 7, 9343. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fotidis, I.A.; Angelidaki, I. Ammonia Effect on Hydrogenotrophic Methanogens and Syntrophic Acetate-Oxidizing Bacteria. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Treu, L.; Kougias, P.G.; De Francisci, D.; Valle, G.; Angelidaki, I. Metagenomic Analysis and Functional Characterization of the Biogas Microbiome Using High Throughput Shotgun Sequencing and a Novel Binning Strategy. Biotechnol. Biofuels 2016, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Kougias, P.G.; Campanaro, S.; Treu, L.; Zhu, X.; Angelidaki, I. “A Novel Archaeal Species Belonging to Methanoculleus Genus Identified via De-Novo Assembly and Metagenomic Binning Process in Biogas Reactors. Anaerobe 2017, 46, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.R.; Fornero, J.J.; Stark, R.; Mets, L.; Angenent, L.T. A Single-Culture Bioprocess of Methanothermobacter Thermautotrophicus to Upgrade Digester Biogas by CO2-to-CH4 Conversion with H2. Archaea 2013, 2013, e157529. [Google Scholar] [CrossRef]
- Treu, L.; Kougias, P.; De Diego-Diaz, B.; Campanaro, S.; Bassani, I.; Fernandez-Rodriguez, J.; Angelidaki, I. Two-Year Microbial Adaptation during Hydrogen-Mediated Biogas Upgrading Process in a Serial Reactor Configuration. Bioresour. Technol. 2018, 264, 140–147. [Google Scholar] [CrossRef]
- Jones, W.J.; Paynter, M.J.B.; Gupta, R. Characterization of Methanococcus Maripaludis Sp. Nov., a New Methanogen Isolated from Salt Marsh Sediment. Arch. Microbiol. 1983, 135, 91–97. [Google Scholar] [CrossRef]
- Dridi, B.; Henry, M.; Khéchine, A.E.; Raoult, D.; Drancourt, M. High Prevalence of Methanobrevibacter Smithii and Methanosphaera Stadtmanae Detected in the Human Gut Using an Improved DNA Detection Protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef]
- Leadbetter, J.R.; Breznak, J.A. Physiological Ecology of Methanobrevibacter Cuticularis Sp. Nov. and Methanobrevibacter Curvatus Sp. Nov., Isolated from the Hindgut of the Termite Reticulitermes Flavipes. Appl. Environ. Microbiol. 1996, 62, 3620–3631. [Google Scholar] [CrossRef]
- Leahy, S.; Kelly, W.J.; Altermann, E.; Ronimus, R.; Yeoman, C.J.; Pacheco, D.M.; Li, N.; Kong, Z.; McTavish, S.; Sang, C.; et al. The Genome Sequence of the Rumen Methanogen Methanobrevibacter Ruminantium Reveals New Possibilities for Controlling Ruminant Methane Emissions. PLoS ONE 2010, 5, e8926. [Google Scholar] [CrossRef]

| Target Genus | Oligo Name * | Oligo Sequence (5′ to 3′) # | Tm [°C] | Length [bp] |
|---|---|---|---|---|
| Methanoculleus (Mcu) a | F3_aEF2_Mcu | TAYCTBATCAACATGATYGAT | 48.0 | 21 |
| FLc_aEF2_Mcu | ACCGCRTCCACSACRAC | 58.4 | 17 | |
| FIP_aEF2_Mcu | GTCTCCGTCTGKGGCATGGTTTTTGACGTGACYCGYGCCATG | 70.9 | 42 | |
| B3_aEF2_Mcu | TYYTCGTTCATDCCCTTGAT | 52.3 | 20 | |
| BL_aEF2_Mcu | ACGAGCAGGAGATGCAGATC | 56.8 | 20 | |
| BIP_aEF2_Mcu | ACCGGCTGRTCAACGAGTTTTTTGACCTTRTCGATCAC | 65.8 | 38 | |
| Methanothermobacter (Mth) b | F3_aEF2_Mth | ATTAAGGAGCTCATGTACCA | 50.6 | 20 |
| FLc_aEF2_Mth | CAAGGAARCGCTGATCCC | 54.9 | 18 | |
| FIP_aEF2_Mth | CCTTGCCTGTTCCTGTTCTTTTGACAACCTCCTGGCTGGTGC | 69.7 | 42 | |
| B3_aEF2_Mth | GCATTATRCCCTCAACKGC | 54.0 | 19 | |
| BL_aEF2_Mth | TCAATGGTCCACTCCTA | 49.1 | 17 | |
| BIP_aEF2_Mth | ACCATTGACGCTGCRAACGTTTTTCCTCATKGCCCTTGTAACGT | 69.1 | 44 | |
| Methanococcus (Mco) c | F3_aEF2_Mco | ATGGGAAGAAGAGCAAAAATG | 51.3 | 21 |
| FLc_aEF2_Mco | ATCATWCCWGCWCCTGC | 52.0 | 17 | |
| FIP_aEF2_Mco | CAAGYTGGTCTCCWGCTTTTCATYGACCACGGTAAAAC | 65.1 | 38 | |
| B3_aEF2_Mco | CTCATTGCTCTTGTAACGTC | 51.1 | 20 | |
| BL_aEF2_Mco | CTGCAAACGTKTCAATGGT | 52.7 | 19 | |
| BIP_aEF2_Mco | GAAGAAGCWGCAAGAGGTATTTTTTCAACGTGACCWGGGGTRTC | 66.4 | 44 | |
| Methanobrevibacter (Mbr) d | F3_aEF2_Mbr | GAAACTGTAYTYAGACAA | 43.5 | 18 |
| FLc_aEF2_Mbr | TCAGGAGCCATGTTYTTGATTA | 53.1 | 22 | |
| FIP_aEF2_Mbr | GCTGAACCRAAWGCTACACTTTTTAATCAACGARTTAAAATTA | 60.6 | 43 | |
| B3_aEF2_Mbr | AAGTGTTCTACTACCATAC | 45.6 | 19 | |
| BL_aEF2_Mbr | ATYATTGATTAYTGTAATG | 39.4 | 19 | |
| BIP_aEF2_Mbr | TGGGCTATYAAYGTTCCTTTTGGTACTTTTTKAGCTAATTC | 61.7 | 41 |
| ID (%) | # Sequences (Uniprot) | # Identical Positions | # Similar Positions | |
|---|---|---|---|---|
| fusA (aEF-2) (K03234) | ||||
| Methanomicrobiales | 61 | 9 | 449 | 157 |
| Methanobacteriales | 47 | 24 | 356 | 192 |
| Methanococcales | 61 | 17 | 447 | 172 |
| mcrA (K00399) | ||||
| Methanomicrobiales | 57 | 15 | 325 | 132 |
| Methanobacteriales | 45 | 41 | 250 | 157 |
| Methanococcales | 66 | 22 | 366 | 117 |
| mer (K00320) | ||||
| Methanomicrobiales | 63 | 9 | 212 | 70 |
| Methanobacteriales | 34 | 24 | 110 | 105 |
| Methanococcales | 58 | 17 | 192 | 75 |
| ehaA (K14097) | ||||
| Methanomicrobiales | 36 | 6 | 63 | 38 |
| Methanobacteriales | 23 | 21 | 48 | 51 |
| Methanococcales | 36 | 17 | 62 | 40 |
| rsp28e (K02979) | ||||
| Methanomicrobiales | 75 | 9 | 52 | 10 |
| Methanobacteriales | 63 | 24 | 46 | 16 |
| Methanococcales | 57 | 17 | 44 | 22 |
| rpl44e (K02929) | ||||
| Methanomicrobiales | 61 | 9 | 56 | 18 |
| Methanobacteriales | 53 | 24 | 50 | 24 |
| Methanococcales | 51 | 17 | 49 | 15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alessi, A.M.; Tao, B.; Zhang, W.; Zhang, Y.; Heaven, S.; Banks, C.J.; Chong, J.P.J. A Rapid, Sensitive, Low-Cost Assay for Detecting Hydrogenotrophic Methanogens in Anaerobic Digesters Using Loop-Mediated Isothermal Amplification. Microorganisms 2020, 8, 740. https://doi.org/10.3390/microorganisms8050740
Alessi AM, Tao B, Zhang W, Zhang Y, Heaven S, Banks CJ, Chong JPJ. A Rapid, Sensitive, Low-Cost Assay for Detecting Hydrogenotrophic Methanogens in Anaerobic Digesters Using Loop-Mediated Isothermal Amplification. Microorganisms. 2020; 8(5):740. https://doi.org/10.3390/microorganisms8050740
Chicago/Turabian StyleAlessi, Anna M., Bing Tao, Wei Zhang, Yue Zhang, Sonia Heaven, Charles J. Banks, and James P. J. Chong. 2020. "A Rapid, Sensitive, Low-Cost Assay for Detecting Hydrogenotrophic Methanogens in Anaerobic Digesters Using Loop-Mediated Isothermal Amplification" Microorganisms 8, no. 5: 740. https://doi.org/10.3390/microorganisms8050740
APA StyleAlessi, A. M., Tao, B., Zhang, W., Zhang, Y., Heaven, S., Banks, C. J., & Chong, J. P. J. (2020). A Rapid, Sensitive, Low-Cost Assay for Detecting Hydrogenotrophic Methanogens in Anaerobic Digesters Using Loop-Mediated Isothermal Amplification. Microorganisms, 8(5), 740. https://doi.org/10.3390/microorganisms8050740







