An Endolysin LysSE24 by Bacteriophage LPSE1 Confers Specific Bactericidal Activity against Multidrug-Resistant Salmonella Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmid and Growth Conditions
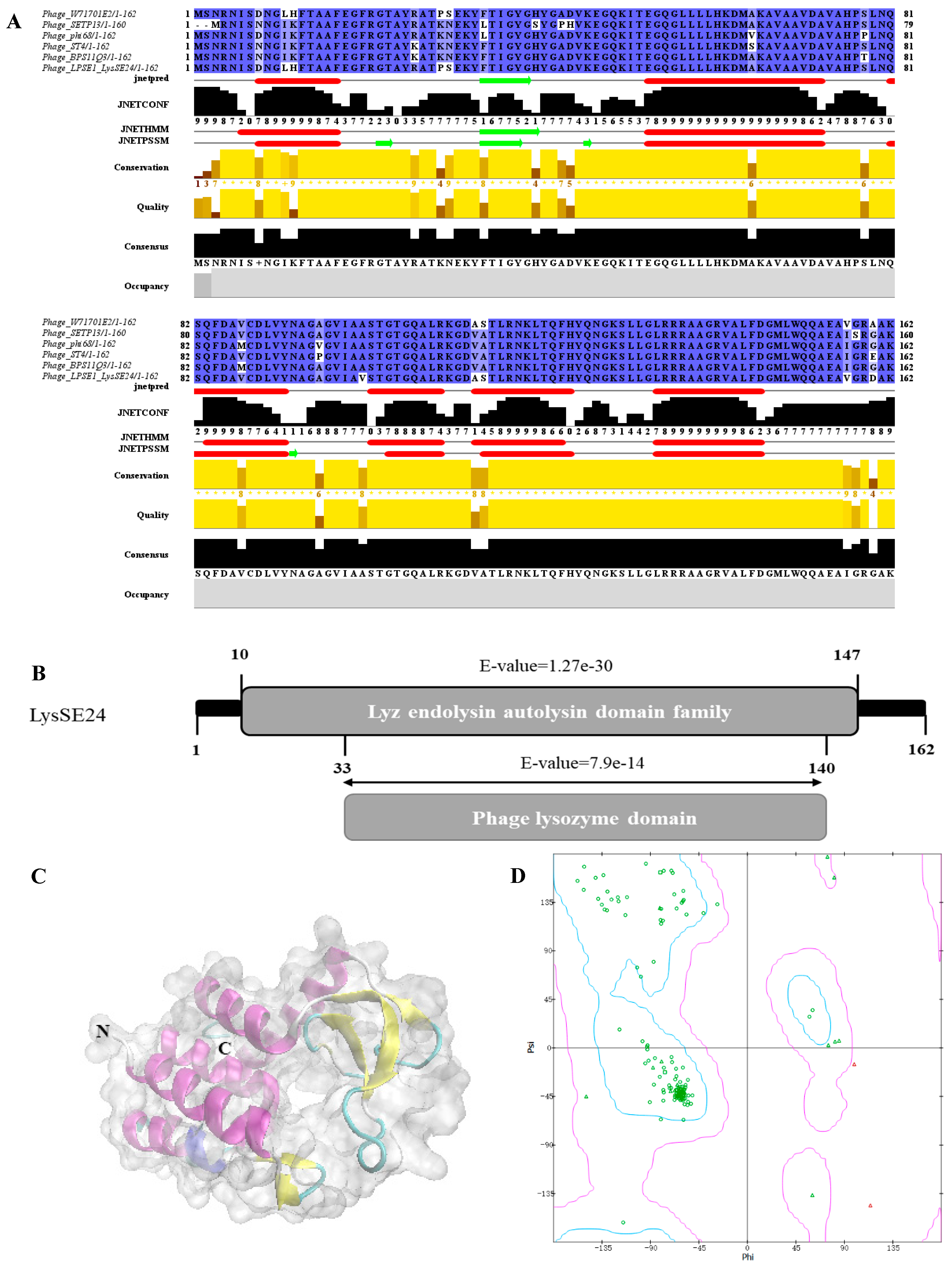
2.2. Bioinformatics Analysis of LysSE24
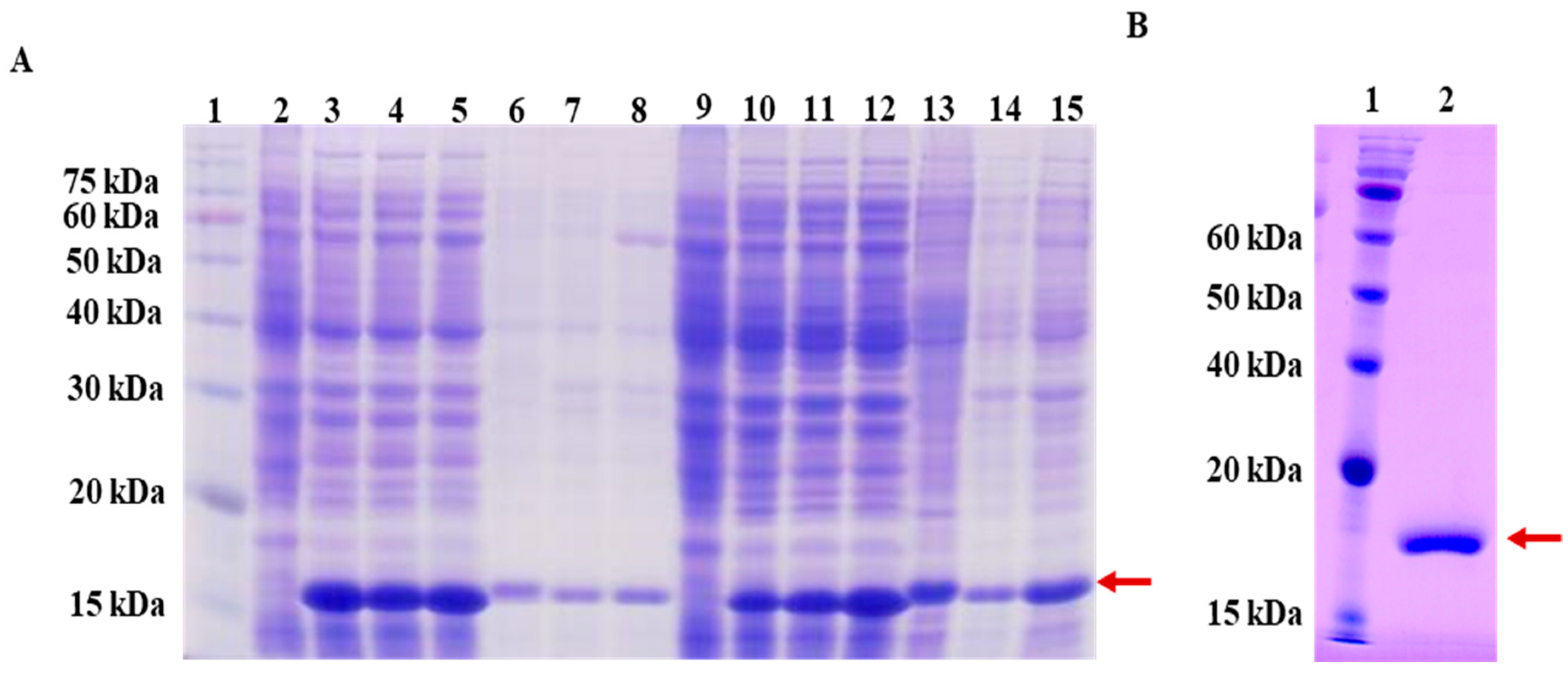
2.3. Cloning, Expression, and Purification of LysSE24
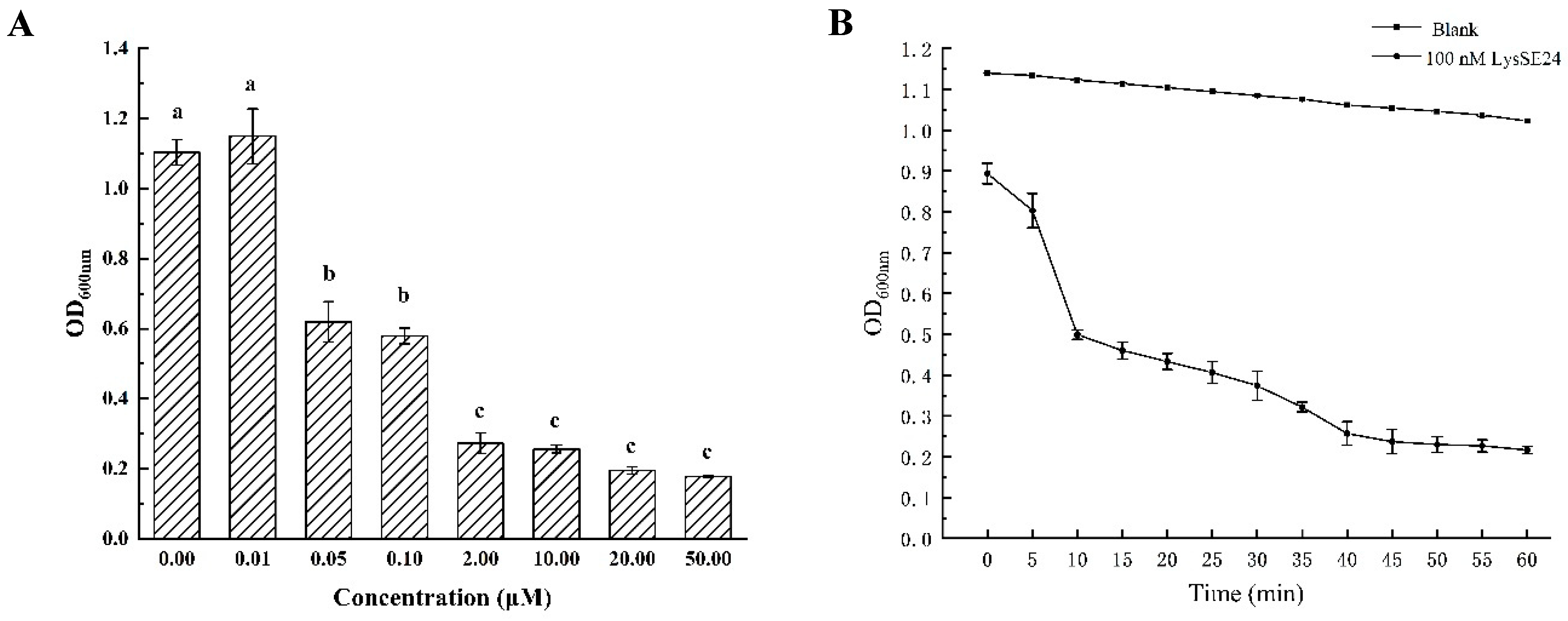
2.4. The Lytic Activity Determination of Endolysin LysSE24
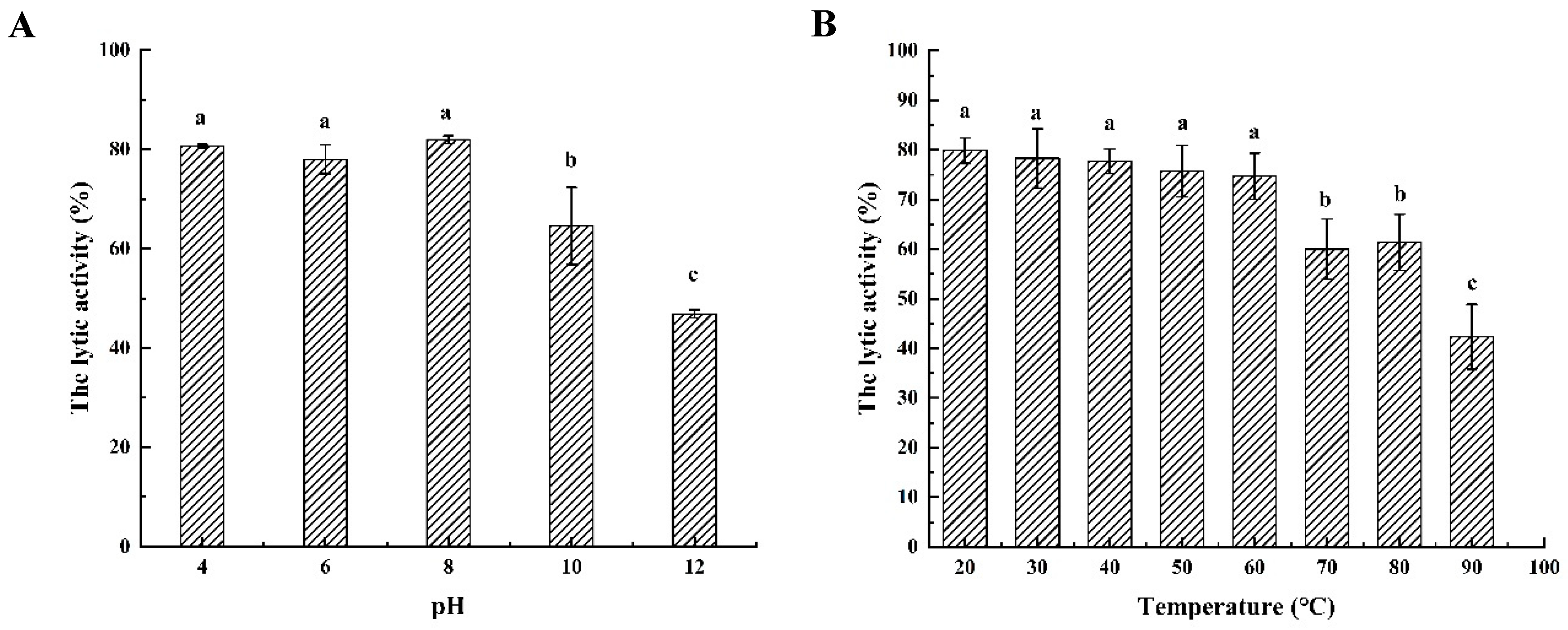
2.5. Biochemical Characterization of Endolysin LysSE24
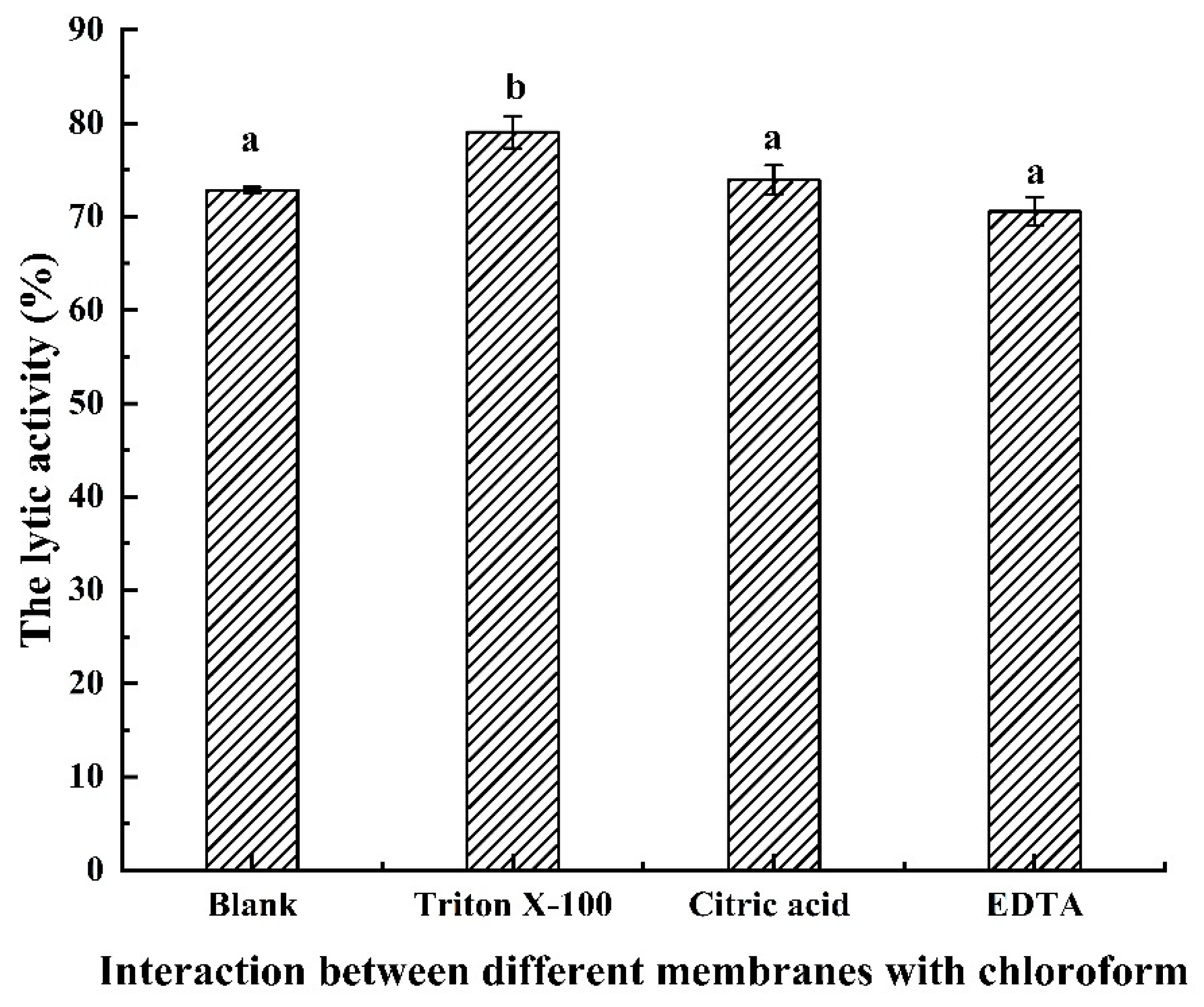
2.6. Antibacterial Activity of LysSE24 in Combination with Outer Membrane Permeabilizers (OMPs)
2.7. The Lytic Spectrum of LysSE24
2.8. Observation by Transmission Electron Microscopic (TEM)
2.9. Statistical Analysis
3. Results and Discussion
3.1. Bioinformatics Analysis of LysSE24
3.2. Large Scale Expression and Purification Endolysin LysSE24
3.3. The Lytic Activity of the Endolysin LysSE24
3.4. Characterization of the Endolysin LysSE24
3.4.1. Temperature Resistance
3.4.2. pH Dependence of Endolysin LysSE24 Enzymatic Activity
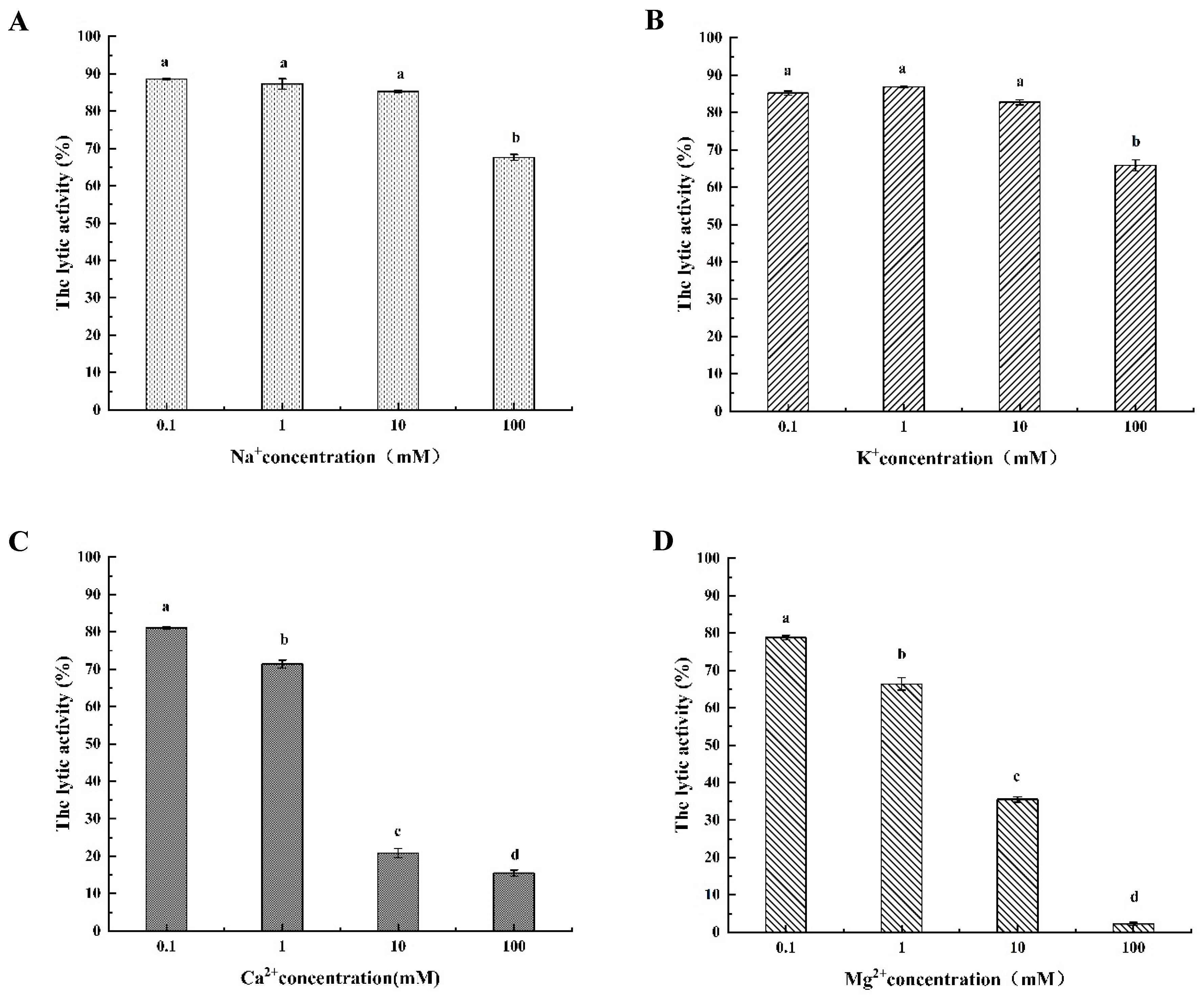
3.4.3. The Different Metal Ionic Effect of LysSE24 Activity
3.5. Antibacterial Activity of LysSE24 in Combination with OMPs
3.6. The Lytic Spectrum of the Endolysin LysSE24
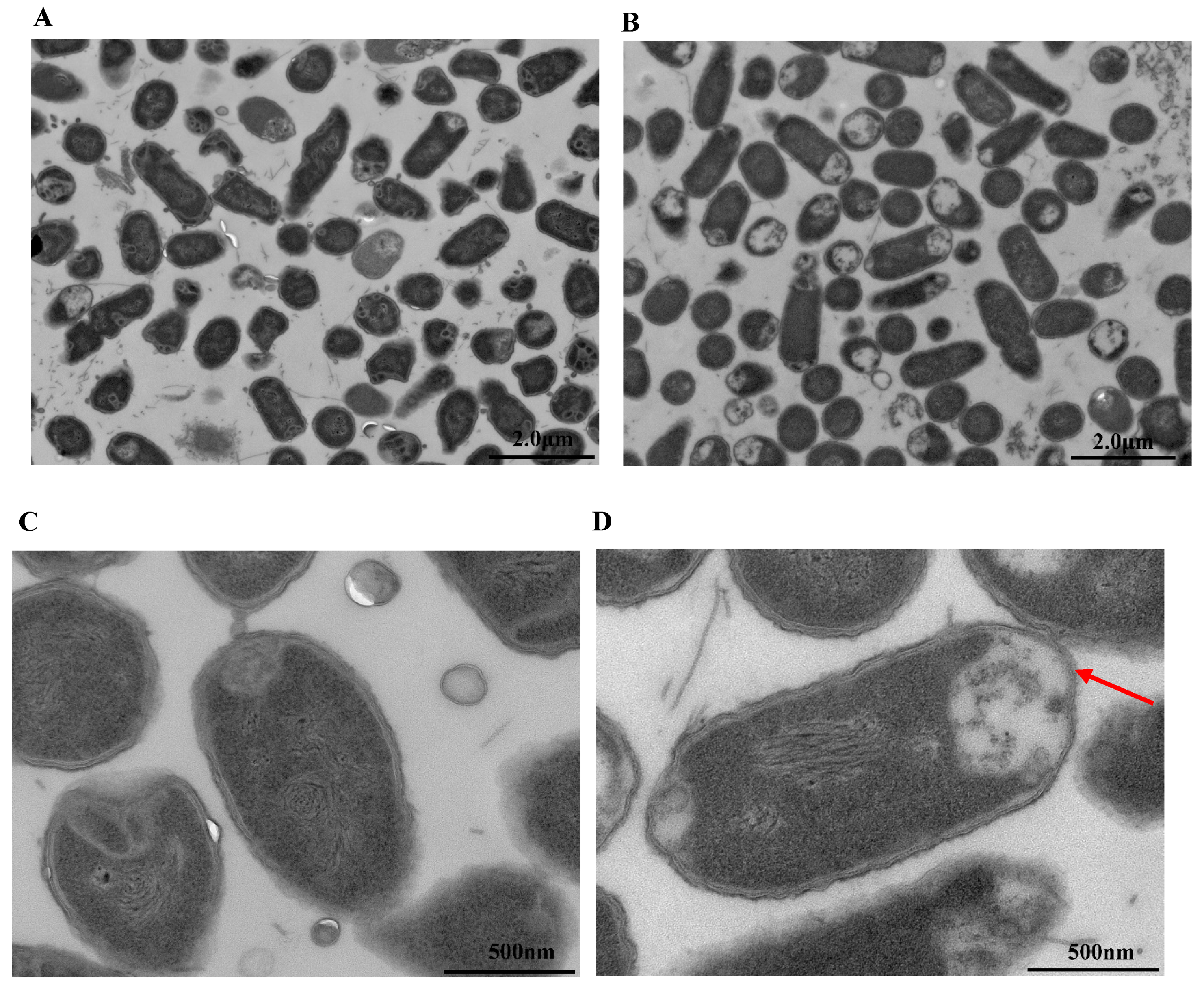
3.7. Observation by Transmission Electron Microscopy (TEM)
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, K.-M.; Runyon, M.; Herrman, T.J.; Phillips, R.; Hsieh, J. Review of Salmonella detection and identification methods: Aspects of rapid emergency response and food safety. Food Control 2015, 47, 264–276. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, 5500. [Google Scholar]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the united states—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Ke, B.; Sun, J.; He, D.; Li, X.; Liang, Z.; Ke, C.-W. Serovar distribution, antimicrobial resistance profiles, and PFGE typing of Salmonella Enterica strains isolated from 2007–2012 in Guangdong, China. BMC Infect. Dis. 2014, 14, 338. [Google Scholar] [CrossRef]
- Luo, Q.; Li, S.; Liu, S.; Tan, H. Foodborne illness outbreaks in China, 2000–2014. Int. J. Clin. Exp. Med. 2017, 10, 5821–5831. [Google Scholar]
- Huang, X.; Huang, Q.; Shi, W.; Liang, J.H.; Ling-Ling, L.U.; Deng, X.L.; Zhang, Y.H. Epidemiological burden of nontyphoidal Salmonella infection in Guangzhou. Chin. J. Food Hyg. 2014, 26, 3. [Google Scholar]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonella interplay with host cells. Nat. Rev. Genet. 2008, 6, 53–66. [Google Scholar] [CrossRef]
- Bula-Rudas, F.J.; Rathore, M.; Maraqa, N.F. Salmonella Infections in Childhood. Adv. Pediatr. 2015, 62, 29–58. [Google Scholar] [CrossRef]
- CDC. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2012 (Final Report); U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2014. [Google Scholar]
- Rohwer, F. Global phage diversity. Cell 2003, 113, 141. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Cao, Z.; Wang, L.; Li, X.; Li, S.; Xu, Y. Bacteriophages as antimicrobial agents against major pathogens in swine: A review. J. Anim. Sci. Biotechnol. 2015, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, M.A.; Oliveira, H.; Azeredo, J. Bacteriophages and Their Role in Food Safety. Int. J. Microbiol. 2012, 2012, 1–13. [Google Scholar] [CrossRef]
- Bai, J.; Kim, Y.-T.; Ryu, S.; Lee, J.-H. Biocontrol and Rapid Detection of Food-Borne Pathogens Using Bacteriophages and Endolysins. Front. Microbiol. 2016, 7, 6230. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage lytic enzymes: Novel anti-infectives. Trends Microbiol. 2005, 13, 491–496. [Google Scholar] [CrossRef]
- Borysowski, J.; Weber-Dabrowska, B.; Górski, A. Bacteriophage Endolysins as a Novel Class of Antibacterial Agents. Exp. Boil. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef]
- Pohane, A.A.; Jain, V. Insights into the regulation of bacteriophage endolysin: Multiple means to the same end. Microbiology 2015, 161, 2269–2276. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins: Applications for food safety. Curr. Opin. Biotechnol. 2016, 37, 76–87. [Google Scholar] [CrossRef]
- Nelson, D.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as Antimicrobials. Adv. Appl. Microbiol. 2012, 83, 299–365. [Google Scholar] [CrossRef]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rubio, L.; Gerstmans, H.; Thorpe, S.; Mesnage, S.; Lavigne, R.; Briers, Y. DUF3380 Domain from a Salmonella Phage Endolysin Shows Potent N-Acetylmuramidase Activity. Appl. Environ. Microbiol. 2016, 82, 4975–4981. [Google Scholar] [CrossRef] [PubMed]
- Díez-Martínez, R.; Fernández, H.D.D.P.; De Paz, H.D.; Bustamante, N.; Euler, C.W.; Fischetti, V.A.; Menéndez, M.; García, P. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J. Antimicrob. Chemother. 2015, 70, 1763–1773. [Google Scholar] [CrossRef]
- Blázquez, B.; Fresco-Taboada, A.; Iglesias-Bexiga, M.; Menéndez, M.; García, P. PL3 Amidase, a Tailor-made Lysin Constructed by Domain Shuffling with Potent Killing Activity against Pneumococci and Related Species. Front. Microbiol. 2016, 7, 2157. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ruiz, I.; Coutinho, F.H.; Rodriguez-Valera, F. Thousands of Novel Endolysins Discovered in Uncultured Phage Genomes. Front. Microbiol. 2018, 9, 1033. [Google Scholar] [CrossRef] [PubMed]
- Larpin, Y.; Oechslin, F.; Moreillon, P.; Resch, G.; Entenza, J.M.; Mancini, S. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS ONE 2018, 13, e0192507. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Liu, J.; Ma, Q.; Zhu, R.; Guo, Z.; Gao, C.; Wang, S.; Yu, L.; Gu, J.; Hu, D.; et al. The N-terminal and central domain of colicin A enables phage lysin to lyse Escherichia coli extracellularly. Antonie Leeuwenhoek 2017, 110, 1627–1635. [Google Scholar] [CrossRef]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, Characterization and Application of Bacteriophage LPSE1 Against Salmonella enterica in Ready to Eat (RTE) Foods. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Mikoulinskaia, G.V.; Odinokova, I.V.; Zimin, A.; Lysanskaya, V.Y.; Feofanov, S.A.; Stepnaya, O.A. Identification and characterization of the metal ion-dependent l-alanoyl-d-glutamate peptidase encoded by bacteriophage T5. FEBS J. 2009, 276, 7329–7342. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Lavigne, R. Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 110, 778–785. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; John, M.W., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Sara, E.-G.; Jaina, M.; Alex, B.; Aurélien, L.; Matloob, Q. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar]
- Drozdetskiy, A.; Cole, C.; Procter, J.B.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar]
- Legotsky, S.A.; Vlasova, K.Y.; Priyma, A.D.; Shneider, M.M.; Pugachev, V.G.; Totmenina, O.D.; Kabanov, A.V.; Miroshnikov, K.; Klyachko, N.L. Peptidoglycan degrading activity of the broad-range Salmonella bacteriophage S-394 recombinant endolysin. Biochimie 2014, 107, 293–299. [Google Scholar] [CrossRef]
- Li, M.; Li, M.; Lin, H.; Wang, J.; Jin, Y.; Han, F. Characterization of the novel T4-like Salmonella Enterica bacteriophage STP4-a and its endolysin. Arch. Virol. 2015, 161, 377–384. [Google Scholar] [CrossRef]
- Lim, J.-A.; Shin, H.; Kang, D.-H.; Ryu, S. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 2012, 163, 233–241. [Google Scholar] [CrossRef]
- Guo, M.; Feng, C.; Ren, J.; Zhuang, X.; Zhang, Y.; Zhu, Y.; Dong, K.; He, P.; Guo, X.; Qin, J. A Novel Antimicrobial Endolysin, LysPA26, against Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 261. [Google Scholar] [CrossRef]
- Geng, P.; Hu, Y.; Zhou, G.; Yuan, Z.; Hu, X. Characterization of three autolysins with activity against cereulide-producing Bacillus isolates in food matrices. Int. J. Food Microbiol. 2017, 241, 291–297. [Google Scholar] [CrossRef]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Cerqueira, M.A.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.; Azeredo, J. A Thermostable Salmonella Phage Endolysin, Lys68, with Broad Bactericidal Properties against Gram-Negative Pathogens in Presence of Weak Acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Powell, A.M.; Camp, M.J.; Pohl, C.S.; Donovan, D.M. Synergistic streptococcal phage lambdaSA2 and B30 endolysins kill Streptococci in cow milk and in a mouse model of mastitis. Appl. Microbiol. Biotechnol. 2015, 99, 8475–8486. [Google Scholar] [CrossRef] [PubMed]
- Jumpei, F.; Tomohiro, N.; Takaaki, F.; Hazuki, O.; Hiromichi, T.; Junya, K.; Masaru, U.; Hidetoshi, H.; Yasunori, T.; Pharmaceuticals, T.Y.J. Characterization of the Lytic Capability of a LysK-Like Endolysin, Lys-phiSA012, Derived from a Polyvalent Staphylococcus aureus Bacteriophage. Pharmaceuticals 2018, 11, 25. [Google Scholar]
- Son, B.; Yun, J.; Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.; Jain, V. Novel method to rapidly and efficiently lyse Escherichia coli for the isolation of recombinant protein. Anal. Biochem. 2017, 528, 1–6. [Google Scholar] [CrossRef]
- Oliveira, H.; Pinto, G.; Oliveira, A.; Oliveira, C.; Faustino, M.A.; Briers, Y.; Domingues, L.; Azeredo, J. Characterization and genome sequencing of a Citrobacter freundii phage CfP1 harboring a lysin active against multidrug-resistant isolates. Appl. Microbiol. Biotechnol. 2016, 100, 10543–10553. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.H.; Lu, C. Purified Recombinant Phage Lysin LySMP: An Extensive Spectrum of Lytic Activity for Swine Streptococci. Curr. Microbiol. 2009, 58, 609–615. [Google Scholar] [CrossRef]
- Pastagia, M.; Schuch, R.; Fischetti, V.A.; Huang, D.B. Lysins: The arrival of pathogen-directed anti-infectives. J. Med Microbiol. 2013, 62, 1506–1516. [Google Scholar] [CrossRef]
- Lu, N.; Sun, Y.; Wang, Q.; Qiu, Y.; Chen, Z.; Wen, Y.; Wang, S.; Song, Y. Cloning and characterization of endolysin and holin from Streptomyces avermitilis bacteriophage phiSASD1 as potential novel antibiotic candidates. Int. J. Boil. Macromol. 2020, 147, 980–989. [Google Scholar] [CrossRef]

| Number | Strain | Strain Type | Source | Drug-Resistance | |
|---|---|---|---|---|---|
| 1 | 13076 | Salmonella Enteritidis | ATCC a | - | |
| 2 | 38 | Salmonella Enteritidis | Lab Collection | + | Amoxycillin, Ampicillin, Nalidixic acid, Sulfonamides |
| 3 | 39 | Salmonella Enteritidis | Lab Collection | + | Amoxycillin, Ampicillin, Nalidixic acid |
| 4 | 42 | Salmonella Enteritidis | Lab Collection | + | Nalidixic acid |
| 5 | 11561 | Salmonella Enteritidis | Lab Collection | + | Ampicillin, Ceftriaxone, Streptomycin, Nalidixic acid, Trimethoprim-sulfamethoxazole, Azithromycin, Fosfomycin |
| 6 | 201 | Salmonella Enteritidis | Lab Collection | + | Ampicillin, Amoxycillin/clavulanic acid, Gentamicin, Spectinomycin, Tetracycline, Sulphafurazole, Apramycin |
| 7 | 211 | Salmonella Enteritidis | Lab Collection | + | Enrofloxacin, Ofloxacin, Apramycin, Colistin |
| 8 | 10960 | Salmonella Enteritidis | Lab Collection | + | Tetracycline, Nalidixic acid, Fosfomycin |
| 9 | 14028 | Salmonella Typhimurium | ATCC | - | |
| 10 | ST-8 | Salmonella Typhimurium | Lab Collection | - | |
| 11 | 30 | Salmonella Typhimurium | Lab Collection | + | Amoxycillin, Ampicillin, Chloramphenicol, Nalidixic acid, Gentamicin, Nitrofurantoin, Sulfonanides, Tetracycline |
| 12 | 36 | Salmonella Typhimurium | Lab Collection | + | Amoxycillin, Ampicillin, Chloramphenicol, Straptomycin, Sulfonanides, Tetracycline |
| 13 | 13336 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Straptomycin, Gentamicin, Kanamycin, Chloramphenicol, Nalidixic acid, Ofloxacin, Ciprofloxacin, Sulphafurazole, Trimethoprim- Sulfamethoxazole, |
| 14 | 13350 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Streptomycin, Gentamicin, Kanamycin, Chloramphenicol, Tetracycline, Nalidixic acid, Ofloxacin, Ciprofloxacin, Sulphafurazole, Trimethoprim-sulfamethoxazole, |
| 15 | 13277 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Streptomycin, Gentamicin, Kanamycin, Chloramphenicol, Tetracycline, Nalidixic acid, Ofloxacin, Ciprofloxacin, Sulphafurazole, Trimethoprim-sulfamethoxazole, |
| 16 | 13337 | Salmonella Typhimurium | Lab Collection | + | Ampicillin Streptomycin, Gentamicin, Kanamycin, Chloramphenicol, Tetracycline, Nalidixic acid, Ofloxacin, Ciprofloxacin, Sulphafurazole, Trimethoprim-sulfamethoxazole, |
| 17 | 13306 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Streptomycin, Gentamicin, Kanamycin, Chloramphenicol, Tetracycline, Nalidixic acid, Ofloxacin, Ciprofloxacin, Sulphafurazole, Trimethoprim-sulfamethoxazole, |
| 18 | 10855 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Ceftriaxone, Ceftiofur, Kanamycin, Streptomycin, Tetracycline, Doxycycline, Ofloxacin, Sulphafurazole |
| 19 | UK-1 | Salmonella Typhimurium | Lab Collection | - | |
| 20 | 114 | Salmonella Typhimurium | Lab Collection | + | Ampicillin, Amoxycillin/Clavulanic acid, Gentamicin, Spectinomycin, Tetracycline, Sulphafurazole, Trimethoprim-sulfamethoxazole, Apramycin, Colistin, Mequindox |
| 21 | 172 | Salmonella Typhimurium | Lab Collection | + | Amoxycillin/Clavulanic acid, Florfenicol, Sulphafurazole, Trimethoprim-sulfamethoxazole, Ceftiofur, Ceftazidime, Meropenem, Apramycin |
| 22 | 206 | Salmonella Typhimurium | Lab Collection | + | Sulphafurazole, Trimethoprim-sulfamethoxazole, Colistin, Mequindox |
| 23 | 17 | Salmonella Argora | Lab Collection | + | Amoxycillin, Ampicillin, Streptomycin, Sulfonamides, Tetracycline |
| 24 | 19 | Salmonella Argora | Lab Collection | + | Amoxycillin, Ampicillin, Cephazolin, Nalidixic acid |
| 25 | 21 | Salmonella Argora | Lab Collection | + | Chloramphenicol |
| 26 | 13500 | Salmonella Indiana | Lab Collection | + | Ampicillin, Amoxycillin/Clavulanic Acid, Streptomycin, Tetracycline, Ceftriaxone, Trimethoprim-sulfamethoxazole, Nalidixic acid, Ciprofloxacin, Fosfomycin, Chloramphenicol, Azithromycin |
| 27 | 13520 | Salmonella Indiana | Lab Collection | + | Ampicillin, Amoxycillin/clavulanic acid, Amikacin, Streptomycin, Tetracycline, Ceftriaxone, Trimethoprim-sulfamethoxazole, Nalidixic acid, Ciprofloxacin, Fosfomycin, Chloramphenicol, Azithromycin |
| 28 | 9270 | Salmonella Anatum | CMCC b | - | |
| 29 | 3723 | Salmonella Dublin | Lab Collection | - | |
| 30 | DH5α | Escherichia coli | Lab Collection | - | |
| 31 | T10 | Escherichia coli | Lab Collection | - | |
| 32 | F18ac | Escherichia coli | Lab Collection | - | |
| 33 | 6538 | Staphylococcus aureus | Lab Collection | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Zhang, Y.; Huang, C.; Wang, J.; Wang, X. An Endolysin LysSE24 by Bacteriophage LPSE1 Confers Specific Bactericidal Activity against Multidrug-Resistant Salmonella Strains. Microorganisms 2020, 8, 737. https://doi.org/10.3390/microorganisms8050737
Ding Y, Zhang Y, Huang C, Wang J, Wang X. An Endolysin LysSE24 by Bacteriophage LPSE1 Confers Specific Bactericidal Activity against Multidrug-Resistant Salmonella Strains. Microorganisms. 2020; 8(5):737. https://doi.org/10.3390/microorganisms8050737
Chicago/Turabian StyleDing, Yifeng, Yu Zhang, Chenxi Huang, Jia Wang, and Xiaohong Wang. 2020. "An Endolysin LysSE24 by Bacteriophage LPSE1 Confers Specific Bactericidal Activity against Multidrug-Resistant Salmonella Strains" Microorganisms 8, no. 5: 737. https://doi.org/10.3390/microorganisms8050737
APA StyleDing, Y., Zhang, Y., Huang, C., Wang, J., & Wang, X. (2020). An Endolysin LysSE24 by Bacteriophage LPSE1 Confers Specific Bactericidal Activity against Multidrug-Resistant Salmonella Strains. Microorganisms, 8(5), 737. https://doi.org/10.3390/microorganisms8050737





