Metagenomic Analysis of Regularly Microwave-Treated and Untreated Domestic Kitchen Sponges
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. DNA Purification
2.3. Library Preparation and Sequencing
2.4. Bioinformatic Analyses
3. Results
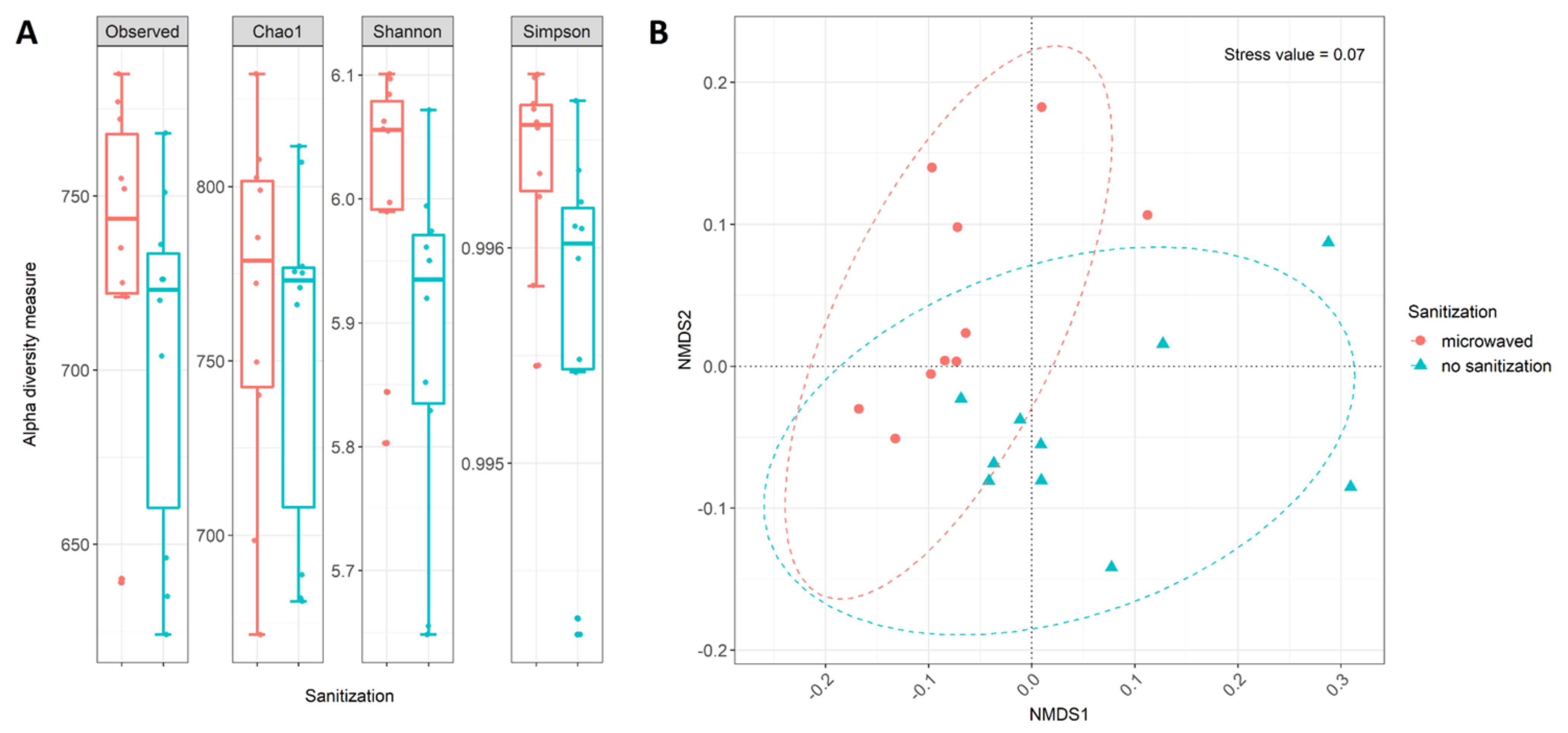
3.1. DNA Extraction and Sequence Analysis
3.2. Taxonomic Differences in Community Composition
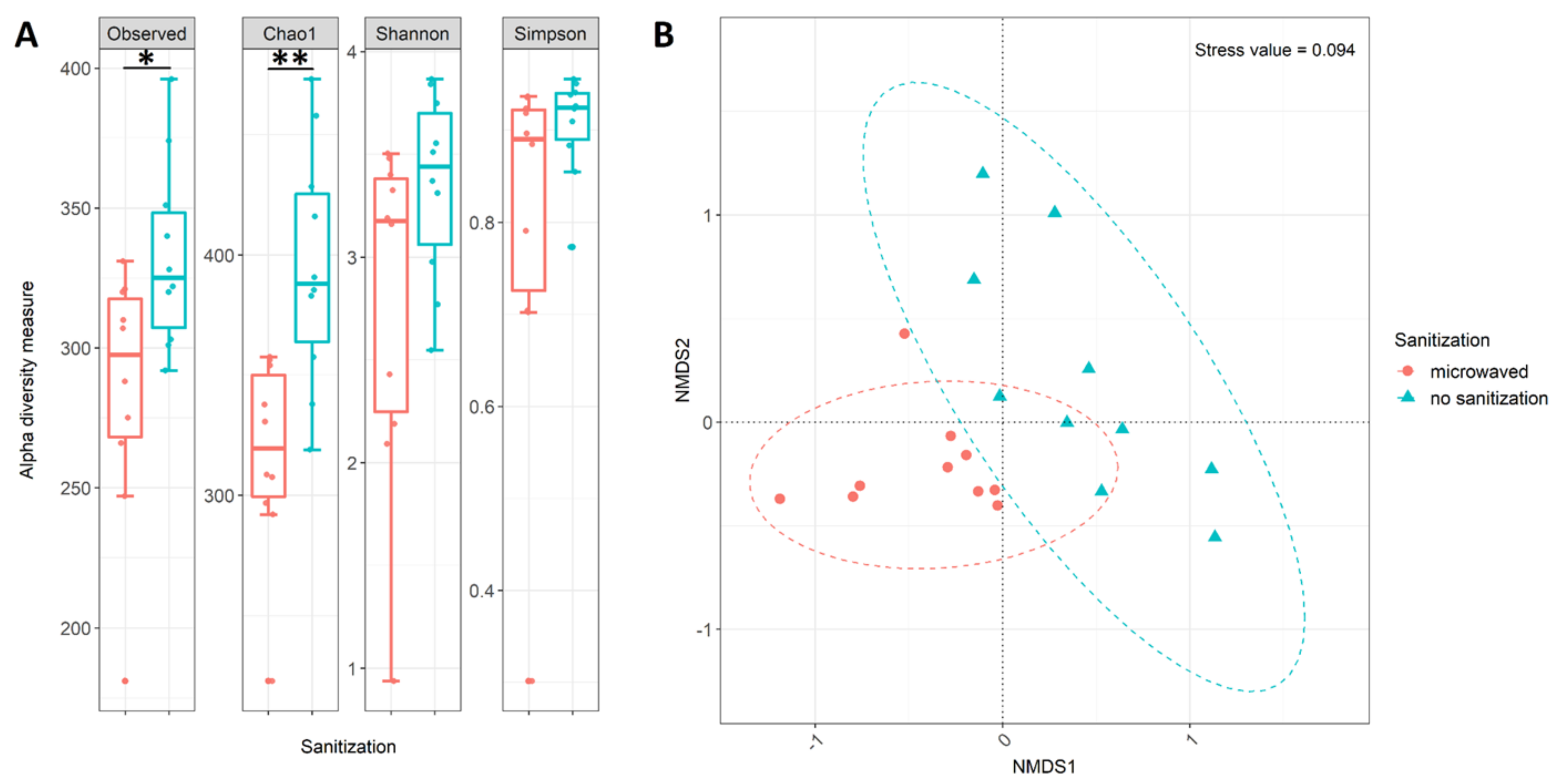
3.3. Differences in Community Structure
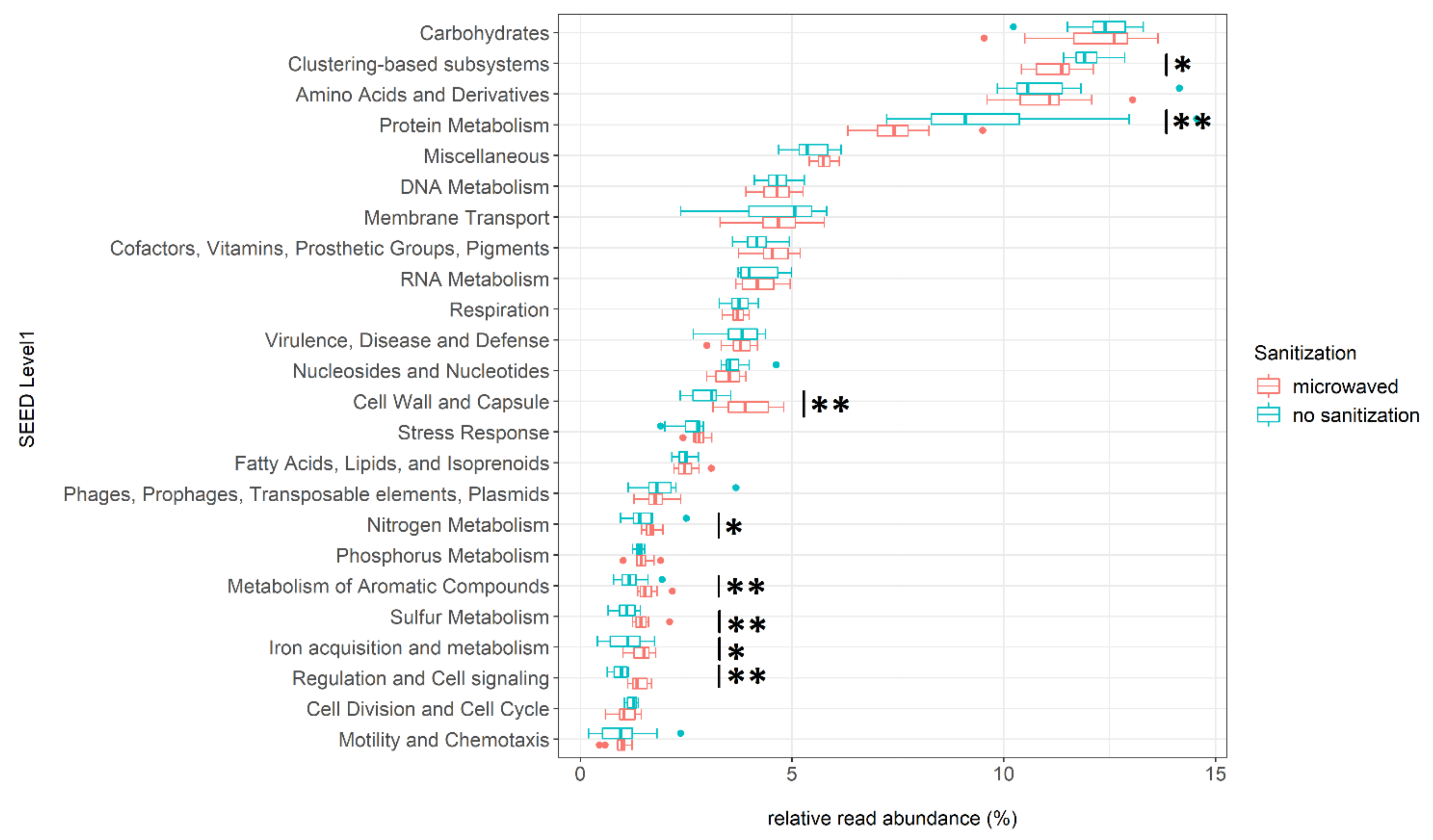
3.4. Differences in Functional Annotation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flores, G.E.; Bates, S.T.; Caporaso, J.G.; Lauber, C.L.; Leff, J.W.; Knight, R.; Fierer, N. Diversity, distribution and sources of bacteria in residential kitchens. Environ Microbiol. Environ. Microbiol. 2013, 15, 588–596. [Google Scholar] [CrossRef]
- Donofrio, R.S.; Bechanko, R.; Hitt, N.; O’Malley, K.; Charnauski, T.; Bestervelt, L.L.; Saha, N.; Saha, R. Are we aware of microbial hotspots in our household? J. Environ. Health 2012, 75, 12–19. [Google Scholar]
- Nielsen, P.; Brumbaugh, E.; Kananen, L. Evaluation of the use of liquid dishwashing compounds to control bacteria in kitchen sponges. J. AOAC Int. 2002, 85, 107–112. [Google Scholar] [CrossRef]
- Gerba, C.P.; Sifuentes, L.Y.; Tamimi, A.H. A Comparison of Urethane and Cellulose Sponges as Cleaning Tools in Household Kitchens. Food Prot. Trends 2017, 37, 170–175. [Google Scholar]
- Wolde, T.; Bacha, K. Microbiological Safety of Kitchen Sponges Used in Food Establishments. Int. J. Food Sci. 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Cardinale, M.; Kaiser, D.; Lueders, T.; Schnell, S.; Egert, M. Microbiome analysis and confocal microscopy of used kitchen sponges reveal massive colonization by Acinetobacter, Moraxella and Chryseobacterium species. Sci. Rep. 2017, 7, 5791. [Google Scholar] [CrossRef]
- Gorman, R.; Bloomfield, S.; Adley, C.C. A study of cross-contamination of food-borne pathogens in the domestic kitchen in the Republic of Ireland. Int. J. Food Microbiol. 2002, 76, 143–150. [Google Scholar] [CrossRef]
- Jovanovska, S.; Quirant, L.; Davidovikj, D.; Popeijus, H.E. Invisible Cohabitants: Investigating the Microbial Presence in the Kitchen Sponges of Maastricht. MJLA 2018, 10, 69–83. [Google Scholar] [CrossRef]
- Marotta, S.M.; Giarratana, F.; Calvagna, A.; Ziino, G.; Giuffrida, A.; Panebianco, A. Study on microbial communities in domestic kitchen sponges: Evidence of Cronobacter sakazakii and Extended Spectrum Beta Lactamase (ESBL) producing bacteria. Ital. J. Food Saf. 2018, 7, 7672. [Google Scholar] [CrossRef]
- Pankaj, K.T.; Shruti, T. Bacterial contamination in kitchens of rural and urban areas in Meerut district of Utter Pradesh (India). Afr. J. Microbiol. Res. 2013, 7, 2020–2026. [Google Scholar] [CrossRef]
- Speirs, J.P.; Anderton, A.; Anderson, J.G. A study of the microbial content of the domestic kitchen. Int. J. Environ. Health Res. 1995, 5, 109–122. [Google Scholar] [CrossRef]
- Taché, J.; Carpentier, B. Hygiene in the home kitchen: Changes in behaviour and impact of key microbiological hazard control measures. Food Control. 2014, 35, 392–400. [Google Scholar] [CrossRef]
- Azevedo, I.; Albano, H.; Silva, J.; Teixeira, P. Food safety in the domestic environment. Food Control. 2014, 37, 272–276. [Google Scholar] [CrossRef]
- Tuladhar, E.; Hazeleger, W.C.; Koopmans, M.; Zwietering, M.H.; Beumer, R.R.; Duizer, E. Residual viral and bacterial contamination of surfaces after cleaning and disinfection. Appl. Environ. Microbiol. 2012, 78, 7769–7775. [Google Scholar] [CrossRef]
- Ikawa, J.Y.; Rossen, J.S. Reducing Bacteria in Household Sponges. Environ Health 1999, 62, 18–22. [Google Scholar]
- Rossi, E.M.; Scapin, D.; Grando, W.F.; Tondo, E.C. Microbiological Contamination and Disinfection Procedures of Kitchen Sponges used in Food Services. FNS 2012, 3, 975–980. [Google Scholar] [CrossRef]
- Sharma, M.; Eastridge, J.; Mudd, C. Effective household disinfection methods of kitchen sponges. Food Control. 2009, 20, 310–313. [Google Scholar] [CrossRef]
- Park, D.K.; Bitton, G.; Melker, R. Microbial inactivation by microwave radiation in the home environment. J. Environ. Health 2006, 69, 17–24. [Google Scholar]
- Shamis, Y.; Croft, R.; Taube, A.; Crawford, R.J.; Ivanova, E.P. Review of the specific effects of microwave radiation on bacterial cells. Appl. Microbiol. Biotechnol. 2012, 96, 319–325. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Jo, E.-K.; Kim, H.-J.; Bai, K.; Park, J.-K. The effects of high-power microwaves on the ultrastructure of Bacillus subtilis. Lett. Appl. Microbiol. 2008, 47, 35–40. [Google Scholar] [CrossRef]
- Shamis, Y.; Taube, A.; Mitik-Dineva, N.; Croft, R.; Crawford, R.J.; Ivanova, E.P. Specific Electromagnetic Effects of Microwave Radiation on Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 3017–3022. [Google Scholar] [CrossRef]
- Woo, I.-S.; Rhee, I.-K.; Park, H.-D. Differential damage in bacterial cells by microwave radiation on the basis of cell wall structure. Appl. Environ. Microbiol. 2000, 66, 2243–2247. [Google Scholar] [CrossRef]
- Janković, S.M.; Milošev, M.Z.; Novaković, M.L. The effects of microwave radiation on microbial cultures. Hosp. Pharm. Int. Multidiscip. J. 2014, 1, 102–108. [Google Scholar] [CrossRef]
- Egert, M. Honor Thy Lodgers?: – Structure and Function of the Human Built Environment Microbiome. TSD 2018, 55, 364–368. [Google Scholar] [CrossRef]
- Gold Biotechnology. Nucleic Acid Precipitation from Dilute Solutions Protocol. Available online: https://www.goldbio.com/documents/3228/Nucleic+Acid+Precipitation.pdf (accessed on 26 November 2019).
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D. vegan: Community Ecology Package. Vegan New 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Manrique, P.; Dills, M.; Young, M.J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef]
- Horne, A.J.; Lessner, D.J. Assessment of the oxidant tolerance of Methanosarcina acetivorans. FEMS MicroBiol. Lett. 2013, 343, 13–19. [Google Scholar] [CrossRef]
- Pavlovsky, L.; Sturtevant, R.A.; Younger, J.G.; Solomon, M.J. Effects of temperature on the morphological, polymeric, and mechanical properties of Staphylococcus epidermidis bacterial biofilms. Langmuir 2015, 31, 2036–2042. [Google Scholar] [CrossRef]
- Olwal, C.O.; Ang’ienda, P.O.; Onyango, D.M.; Ochiel, D.O. Susceptibility patterns and the role of extracellular DNA in Staphylococcus epidermidis biofilm resistance to physico-chemical stress exposure. BMC Microbiol. 2018, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Kent, A.G.; Garcia, C.A.; Martiny, A.C. Increased biofilm formation due to high-temperature adaptation in marine Roseobacter. Nat. Microbiol. 2018, 3, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.K.; Rainey, P.B.; Haagensen, J.A.; Molin, S. Evolution of species interactions in a biofilm community. Nature 2007, 445, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Mohan, R.; Gupta, R.S. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2009, 59, 234–247. [Google Scholar] [CrossRef]
- Rabus, R.; Hansen, T.A.; Widdel, F. Dissimilatory Sulfate- and Sulfur-Reducing Prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 309–404. [Google Scholar] [CrossRef]
- Xu, K.D.; Stewart, P.S.; Xia, F.; Huang, C.T.; McFeters, G.A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 1998, 64, 4035–4039. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef]
| Taxonomy Level | Organism | Microwaved (%) | No Treatment (%) | p-Values |
|---|---|---|---|---|
| Phylum | Bacteroidetes | 2.0 | 12.9 | 0.009 |
| Proteobacteria | 92.0 | 79.2 | 0.013 | |
| Class | Sphingobacteriia | 0.2 | 1.2 | 0.008 |
| Flavobacteriia | 1.6 | 11.2 | 0.009 | |
| Gammaproteobacteria | 71.3 | 33.6 | 0.009 | |
| Betaproteobacteria | 2.7 | 7.5 | 0.031 | |
| Order | Sphingobacteriales | 0.2 | 1.2 | 0.006 |
| Flavobacteriales | 1.5 | 11.2 | 0.011 | |
| Aeromonadales | 2.0 | 0.2 | 0.006 | |
| Enterobacteriales | 19.2 | 3.7 | 0.006 | |
| Pseudomonadales | 48.1 | 24.1 | 0.035 | |
| Burkholderiales | 2.4 | 7.2 | 0.035 | |
| Family | Sphingobacteriaceae | 0.2 | 1.1 | 0.011 |
| Flavobacteriaceae | 1.5 | 11.0 | 0.018 | |
| Aeromonadaceae | 2.0 | 0.2 | 0.011 | |
| Enterobacteriaceae | 19.2 | 3.7 | 0.011 | |
| Moraxellaceae | 40.6 | 21.7 | 0.046 | |
| Pseudomonadaceae | 7.6 | 2.4 | 0.023 | |
| Alcaligenaceae | 0.3 | 3.6 | 0.023 | |
| Brucellaceae | 0.6 | 4.9 | 0.031 | |
| Genus | Chryseobacterium | 0.5 | 3.6 | 0.011 |
| Riemerella | 0.2 | 1.1 | 0.015 | |
| unclassified (Flavobacteriaceae) | 0.3 | 4.8 | 0.011 | |
| Aeromonas | 1.9 | 0.2 | 0.011 | |
| Citrobacter | 2.7 | 0.3 | 0.011 | |
| Salmonella | 1.4 | 0.3 | 0.011 | |
| Escherichia | 2.0 | 0.4 | 0.011 | |
| Enterobacter | 5.8 | 0.9 | 0.011 | |
| Klebsiella | 5.7 | 1.4 | 0.018 | |
| Acinetobacter | 34.6 | 8.4 | 0.011 | |
| Pseudomonas | 7.4 | 2.4 | 0.020 | |
| Bordetella | 0.1 | 1.0 | 0.011 | |
| Achromobacter | 0.2 | 2.6 | 0.024 | |
| Brucella | 0.2 | 2.0 | 0.024 | |
| Ochrobactrum | 0.5 | 2.9 | 0.033 | |
| Caulobacter | 0.4 | 4.4 | 0.043 |
| Kingdom | Eukaryota | Archaea | Viruses | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxonomy Level | Organism | Microwaved (%) | No Treatment (%) | Sig. | Organism | Microwaved (%) | No Treatment (%) | Sig. | Organism | Microwaved (%) | NoTreatment (%) | Sig. |
| Class | unclassified (Eukaryota) | 0.000 | 0.004 | * | none | - | - | - | none | - | - | - |
| Order | unclassified (Eukaryota) | 0.000 | 0.004 | * | Methanosarcinales | 0.008 | 0.000 | * | Caudovirales | 0.064 | 0.025 | * |
| Family | unclassified (Eukaryota) | 0.000 | 0.004 | * | Methanosarcinaceae | 0.008 | 0.000 | * | Myoviridae | 0.028 | 0.005 | * |
| Metschnikowiaceae | 0.003 | 0.000 | * | Podoviridae | 0.010 | 0.001 | * | |||||
| Siphoviridae | 0.027 | 0.018 | * | |||||||||
| Genus | Acanthamoeba | 0.000 | 0.004 | * | Methanosarcina | 0.005 | 0.000 | * | Lambda-like viruses | 0.008 | 0.003 | * |
| Clavispora | 0.003 | 0.000 | * | P2-like viruses | 0.027 | 0.002 | * | |||||
| Aspergillus | 0.000 | 0.004 | * | T7-like viruses | 0.006 | 0.001 | * | |||||
| unclassified (Podoviridae) | 0.003 | 0.000 | * | |||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacksch, S.; Thota, J.; Shetty, S.; Smidt, H.; Schnell, S.; Egert, M. Metagenomic Analysis of Regularly Microwave-Treated and Untreated Domestic Kitchen Sponges. Microorganisms 2020, 8, 736. https://doi.org/10.3390/microorganisms8050736
Jacksch S, Thota J, Shetty S, Smidt H, Schnell S, Egert M. Metagenomic Analysis of Regularly Microwave-Treated and Untreated Domestic Kitchen Sponges. Microorganisms. 2020; 8(5):736. https://doi.org/10.3390/microorganisms8050736
Chicago/Turabian StyleJacksch, Susanne, Jyothi Thota, Sudarshan Shetty, Hauke Smidt, Sylvia Schnell, and Markus Egert. 2020. "Metagenomic Analysis of Regularly Microwave-Treated and Untreated Domestic Kitchen Sponges" Microorganisms 8, no. 5: 736. https://doi.org/10.3390/microorganisms8050736
APA StyleJacksch, S., Thota, J., Shetty, S., Smidt, H., Schnell, S., & Egert, M. (2020). Metagenomic Analysis of Regularly Microwave-Treated and Untreated Domestic Kitchen Sponges. Microorganisms, 8(5), 736. https://doi.org/10.3390/microorganisms8050736






