Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases
Abstract
1. Introduction
2. Methods
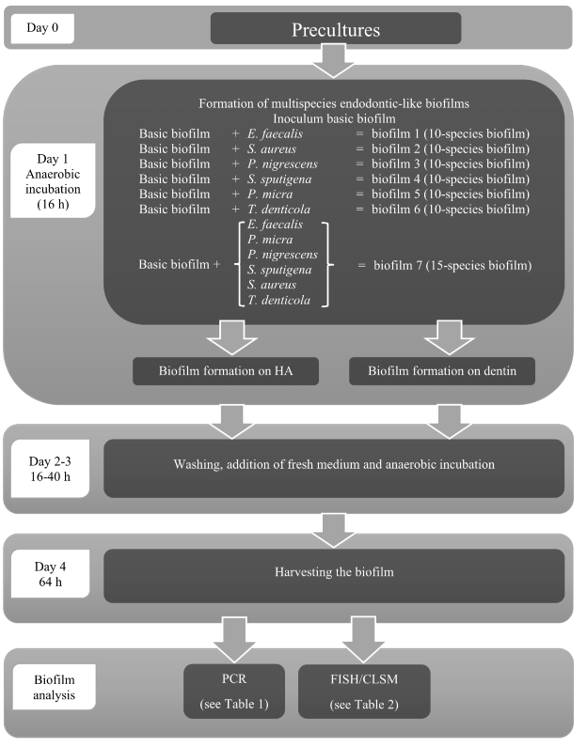
2.1. Multispecies Endodontic-Like Biofilm Formation
2.2. Biofilm Quantification Using Quantitative Real-Time PCR (qPCR)
2.3. Fluorescence in Situ Hybridization (FISH)
2.4. Confocal Laser Scanning Microscopy (CLSM)
2.5. Statistical Analysis
3. Results
3.1. The Addition of Endodontic Pathogens Induced Significant Changes in Cell Counts within Endodontic-Like Biofilms on HA
3.2. The Bacterial Composition of Endodontic-Like Biofilms on Dentin Was Also Substantially Affected by the Presence of Endodontic Pathogens
3.3. Different Substrates Did Not Affect the Composition of the Endodontic-Like Multispecies Biofilms
3.4. FISH/CLSM Reveals E. faecalis Aggregates and S. aureus Microcolonies within Endodontic-Like Biofilms
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Henderson, B.; Wilson, M. Commensal communism and the oral cavity. J. Dent. Res. 1998, 77, 1674–1683. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Jhajharia, K.; Parolia, A.; Shetty, K.V.; Mehta, L.K. Biofilm in endodontics: A review. J. Int. Soc. Prev. Community Dent. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Ali, I.A.A.; Cheung, B.P.K.; Yau, J.Y.Y.; Matinlinna, J.P.; Levesque, C.M.; Belibasakis, G.N.; Neelakantan, P. The influence of substrate surface conditioning and biofilm age on the composition of Enterococcus faecalis biofilms. Int. Endod. J. 2020, 53, 53–61. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020. [Google Scholar] [CrossRef]
- Flemming, H.C. Biofouling and me: My Stockholm syndrome with biofilms. Water Res. 2020, 173, 115576. [Google Scholar] [CrossRef]
- Kolenbrander, P.; Palmer, R.J.; Periasamy, S.; Jakubovics, N. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Larsen, T.; Fiehn, N.E. Dental biofilm infections—An update. Apmis 2017, 125, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, L.L.; Vaishnavi, C. Endodontic microbiology. J. Conserv Dent. 2010, 13, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Taschieri, S.; Del Fabbro, M.; Samaranayake, L.; Chang, J.W.; Corbella, S. Microbial invasion of dentinal tubules: A literature review and a new perspective. J. Investig Clin. Dent. 2014, 5, 163–170. [Google Scholar] [CrossRef]
- Fouad, A.F. Endodontic Microbiology and Pathobiology: Current State of Knowledge. Dent. Clin. N. Am. 2017, 61, 1–15. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Palazzi, F.; Giardino, L.; Shalavi, S. Microbial biofilms in endodontic infections: An update review. Biomed. J. 2013, 36, 59–70. [Google Scholar] [CrossRef]
- Nair, P.N.R. Light and electron microscopic studies of root canal flora and periapical lesions. J. Endod. 1987, 13, 29–39. [Google Scholar] [CrossRef]
- Ricucci, D.; Candeiro, G.T.; Bugea, C.; Siqueira, J.F., Jr. Complex Apical Intraradicular Infection and Extraradicular Mineralized Biofilms as the Cause of Wet Canals and Treatment Failure: Report of 2 Cases. J. Endod. 2016, 42, 509–515. [Google Scholar] [CrossRef]
- Ricucci, D.; Siqueira, J.F., Jr. Biofilms and apical periodontitis: Study of prevalence and association with clinical and histopathologic findings. J. Endod. 2010, 36, 1277–1288. [Google Scholar] [CrossRef]
- Neelakantan, P.; Romero, M.; Vera, J.; Daood, U.; Khan, A.U.; Yan, A.; Cheung, G.S.P. Biofilms in Endodontics-Current Status and Future Directions. Int. J. Mol. Sci. 2017, 18, 1748. [Google Scholar] [CrossRef]
- Sakamoto, M.; Siqueira, J.F., Jr.; Rocas, I.N.; Benno, Y. Molecular analysis of the root canal microbiota associated with endodontic treatment failures. Oral Microbiol. Immunol. 2008, 23, 275–281. [Google Scholar] [CrossRef]
- Gomes, B.P.; Pinheiro, E.T.; Gade-Neto, C.R.; Sousa, E.L.; Ferraz, C.C.; Zaia, A.A.; Teixeira, F.B.; Souza-Filho, F.J. Microbiological examination of infected dental root canals. Oral Microbiol. Immunol. 2004, 19, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ammann, T.W.; Belibasakis, G.N.; Thurnheer, T. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS ONE 2013, 8 12, e83090. [Google Scholar] [CrossRef]
- Ammann, T.; Gmur, R.; Thurnheer, T. Advancement of the 10-species subgingival Zurich Biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol. 2012, 12, 227. [Google Scholar] [CrossRef] [PubMed]
- Guggenheim, B.; Gmur, R.; Galicia, J.; Stathopoulou, P.; Benakanakere, M.; Meier, A.; Thurnheer, T.; Kinane, D. In vitro modeling of host-parasite interactions: The ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009, 9, 280. [Google Scholar] [CrossRef]
- Guggenheim, B.; Giertsen, E.; Schüpbach, P.; Shapiro, S. Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 2001, 80, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.; Thurnheer, T. Validation of antibiotic efficacy on in vitro subgingival biofilms. J. Periodontol. 2014, 85, 343–348. [Google Scholar] [CrossRef]
- Thurnheer, T.; Belibasakis, G.; Bostanci, N. Colonisation of gingival epithelia by subgingival biofilms in vitro: Role of “red complex” bacteria. Arch. Oral Biol. 2014, 59, 977–986. [Google Scholar] [CrossRef]
- Munson, M.A.; Pitt-Ford, T.; Chong, B.; Weightman, A.; Wade, W.G. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 2002, 81, 761–766. [Google Scholar] [CrossRef]
- Rolph, H.J.; Lennon, A.; Riggio, M.P.; Saunders, W.P.; MacKenzie, D.; Coldero, L.; Bagg, J. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 2001, 39, 3282–3289. [Google Scholar] [CrossRef]
- Tomazinho, L.F.; Avila-Campos, M.J. Detection of Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia, and Prevotella nigrescens in chronic endodontic infection. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007, 103, 285–288. [Google Scholar] [CrossRef]
- Ammann, T.; Bostanci, N.; Belibasakis, G.; Thurnheer, T. Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model. J. Periodontal. Res. 2013, 48, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Belibasakis, G. Incorporation of staphylococci into titanium-grown biofilms: An in vitro “submucosal” biofilm model for peri-implantitis. Clin. Oral Implant. Res. 2016, 27, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Gmür, R.; Guggenheim, B. Antigenic heterogeneity of Bacteroides intermedius as recognized by monoclonal antibodies. Infect. Immun. 1983, 42, 459–470. [Google Scholar] [CrossRef]
- Sedgley, C.M.; Nagel, A.C.; Shelburne, C.E.; Clewell, D.B.; Appelbe, O.; Molander, A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch. Oral Biol. 2005, 50, 575–583. [Google Scholar] [CrossRef]
- Rechenberg, D.; Thurnheer, T.; Zehnder, M. Potential systematic error in laboratory experiments on microbial leakage through filled root canals: An experimental study. Int. Endod. J. 2011, 44, 827–835. [Google Scholar] [CrossRef]
- Thurnheer, T.; Gmür, R.; Guggenheim, B. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 2004, 56, 37–47. [Google Scholar] [CrossRef]
- Gmür, R.; Thurnheer, T. Direct quantitative differentiation between Prevotella intermedia and Prevotella nigrescens in clinical specimens. Microbiology 2002, 148, 1379–1387. [Google Scholar] [CrossRef][Green Version]
- Zehnder, M.; Rechenberg, D.; Thurnheer, T.; Lüthi-Schaller, H.; Belibasakis, G. FISHing for gutta-percha-adhered biofilms in purulent post-treatment apical periodontitis. Mol. Oral Microbiol. 2017, 32, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Zijnge, V.; van Leeuwen, M.; Degener, J.; Abbas, F.; Thurnheer, T.; Gmur, R.; Harmsen, H. Oral biofilm architecture on natural teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef]
- Thurnheer, T.; Belibasakis, G. Integration of non-oral bacteria into in vitro oral biofilms. Virulence 2015, 6, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Ohashi, A.; Imachi, H.; Harada, H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol. 2006, 72, 5311–5317. [Google Scholar] [CrossRef]
- Brosius, J.; Dull, T.J.; Sleeter, D.D.; Noller, H.F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 1981, 148, 107–127. [Google Scholar] [CrossRef]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Demuth, D. Quorum sensing regulation of biofilm growth and gene expression by oral bacteria and periodontal pathogens. Periodontology 2000 2010, 52, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Reading, N.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Yamada, M.; Ikegami, A.; Kuramitsu, H.K. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett. 2005, 250, 271–277. [Google Scholar] [CrossRef]
- Kreth, J.; Merritt, J.; Shi, W.; Qi, F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 2005, 187, 7193–7203. [Google Scholar] [CrossRef]
- Shapiro, S.; Giertsen, E.; Guggenheim, B. An in vitro oral biofilm model for comparing the efficacy of antimicrobial mouthrinses. Caries Res. 2002, 36, 93–100. [Google Scholar] [CrossRef]
- Guggenheim, B.; Guggenheim, M.; Gmür, R.; Giertsen, E.; Thurnheer, T. Application of the Zürich biofilm model to problems of cariology. Caries Res. 2004, 38, 212–222. [Google Scholar] [CrossRef]
- Guggenheim, M.; Shapiro, S.; Gmür, R.; Guggenheim, B. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 2001, 67, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Belibasakis, G. Effect of sodium fluoride on oral biofilm microbiota and enamel demineralization. Arch. Oral Biol. 2018, 89, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Bostanci, N.; Belibasakis, G. Microbial dynamics during conversion from supragingival to subgingival biofilms in an in vitro model. Mol. Oral Microbiol. 2016, 31, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Karygianni, L.; Flury, M.; Belibasakis, G.N. Fusobacterium Species and Subspecies Differentially Affect the Composition and Architecture of Supra- and Subgingival Biofilms Models. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Jung, D.J.; Al-Ahmad, A.; Follo, M.; Spitzmuller, B.; Hoth-Hannig, W.; Hannig, M.; Hannig, C. Visualization of initial bacterial colonization on dentine and enamel in situ. J. Microbiol. Methods 2010, 81, 166–174. [Google Scholar] [CrossRef]
- Rocas, I.N.; Siqueira, J.F., Jr.; Santos, K.R. Association of Enterococcus faecalis with different forms of periradicular diseases. J. Endod. 2004, 30, 315–320. [Google Scholar] [CrossRef]
- Dioguardi, M.; Di Gioia, G.; Illuzzi, G.; Arena, C.; Caponio, V.C.A.; Caloro, G.A.; Zhurakivska, K.; Adipietro, I.; Troiano, G.; Lo Muzio, L. Inspection of the Microbiota in Endodontic Lesions. Dent. J. Basel 2019, 7, 47. [Google Scholar] [CrossRef]
- Chavez de Paz, L.E. Redefining the persistent infection in root canals: Possible role of biofilm communities. J. Endod. 2007, 33, 652–662. [Google Scholar] [CrossRef]
- Ponce, J.B.; Midena, R.Z.; Pinke, K.H.; Weckwerth, P.H.; Andrade, F.B.; Lara, V.S. In vitro treatment of Enterococcus faecalis with calcium hydroxide impairs phagocytosis by human macrophages. Acta Odontol. Scand. 2019, 77, 158–163. [Google Scholar] [CrossRef]
- Distel, J.W.; Hatton, J.F.; Gillespie, M.J. Biofilm formation in medicated root canals. J. Endod. 2002, 28, 689–693. [Google Scholar] [CrossRef]
- Sundqvist, G.; Figdor, D.; Persson, S.; Sjogren, U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1998, 85, 86–93. [Google Scholar] [CrossRef]
- Chavez de Paz, L.E.; Davies, J.R.; Bergenholtz, G.; Svensater, G. Strains of Enterococcus faecalis differ in their ability to coexist in biofilms with other root canal bacteria. Int. Endod. J. 2015, 48, 916–925. [Google Scholar] [CrossRef]
- Ran, S.J.; Jiang, W.; Zhu, C.L.; Liang, J.P. Exploration of the mechanisms of biofilm formation by Enterococcus faecalis in glucose starvation environments. Aust. Dent. J. 2015, 60, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Jacinto, R.C.; Gomes, B.P.; Desai, M.; Rajendram, D.; Shah, H.N. Bacterial examination of endodontic infections by clonal analysis in concert with denaturing high-performance liquid chromatography. Oral Microbiol. Immunol. 2007, 22, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Vieira Colombo, A.P.; Magalhaes, C.B.; Hartenbach, F.A.; Martins do Souto, R.; Maciel da Silva-Boghossian, C. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb. Pathog. 2016, 94, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Makovcova, J.; Babak, V.; Kulich, P.; Masek, J.; Slany, M.; Cincarova, L. Dynamics of mono- and dual-species biofilm formation and interactions between Staphylococcus aureus and Gram-negative bacteria. Microb. Biotechnol. 2017, 10, 819–832. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hebraud, M.; Moretro, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.J.; Kacaniova, M.; Czaczyk, K.; et al. Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 2015, 6, 841. [Google Scholar] [CrossRef]
- Bae, K.S.; Baumgartner, J.C.; Shearer, T.R.; David, L.L. Occurrence of Prevotella nigrescens and Prevotella intermedia in infections of endodontic origin. J. Endod. 1997, 23, 620–623. [Google Scholar] [CrossRef]
- Gharbia, S.E.; Haapasalo, M.; Shah, H.N.; Kotiranta, A.; Lounatmaa, K.; Pearce, M.A.; Devine, D.A. Characterization of Prevotella intermedia and Prevotella nigrescens isolates from periodontic and endodontic infections. J. Periodontol. 1994, 65, 56–61. [Google Scholar] [CrossRef]
- Gomes, B.P.; Jacinto, R.C.; Pinheiro, E.T.; Sousa, E.L.; Zaia, A.A.; Ferraz, C.C.; Souza-Filho, F.J. Porphyromonas gingivalis, Porphyromonas endodontalis, Prevotella intermedia and Prevotella nigrescens in endodontic lesions detected by culture and by PCR. Oral Microbiol. Immunol. 2005, 20, 211–215. [Google Scholar] [CrossRef]
- Saito, K.; Takahashi, N.; Horiuchi, H.; Yamada, T. Effects of glucose on formation of cytotoxic end-products and proteolytic activity of Prevotella intermedia, Prevotella nigrescens and Porphyromonas gingivalis. J. Periodontal. Res. 2001, 36, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Dashper, S.G.; Ang, C.S.; Veith, P.D.; Mitchell, H.L.; Lo, A.W.; Seers, C.A.; Walsh, K.A.; Slakeski, N.; Chen, D.; Lissel, J.P.; et al. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J. Bacteriol. 2009, 191, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Saito, K.; Schachtele, C.F.; Yamada, T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol. Immunol. 1997, 12, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Rocas, I.N.; Siqueira, J.F., Jr.; Debelian, G.J. Analysis of symptomatic and asymptomatic primary root canal infections in adult Norwegian patients. J. Endod. 2011, 37, 1206–1212. [Google Scholar] [CrossRef]
- Zhu, Y.; Dashper, S.; Chen, Y.; Crawford, S.; Slakeski, N.; Reynolds, E. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS ONE 2013, 8, e71727. [Google Scholar] [CrossRef]
- Sanghavi, T.H.; Shah, N.; Shah, R.R.; Sanghavi, A. Investigate the correlation between clinical sign and symptoms and the presence of P. gingivalis, T. denticola, and T. forsythia individually or as a “Red complex” by a multiplex PCR method. J. Conserv. Dent. 2014, 17, 555–560. [Google Scholar] [CrossRef]
- Deng, Z.L.; Sztajer, H.; Jarek, M.; Bhuju, S.; Wagner-Dobler, I. Worlds Apart—Transcriptome Profiles of Key Oral Microbes in the Periodontal Pocket Compared to Single Laboratory Culture Reflect Synergistic Interactions. Front. Microbiol. 2018, 9, 124. [Google Scholar] [CrossRef]
- Neilands, J.; Davies, J.R.; Bikker, F.J.; Svensater, G. Parvimonas micra stimulates expression of gingipains from Porphyromonas gingivalis in multi-species communities. Anaerobe 2019, 55, 54–60. [Google Scholar] [CrossRef]
- Chavez de Paz, L.; Hamilton, I.; Svensater, G. Oral bacteria in biofilms exhibit slow reactivation from nutrient deprivation. Microbiology 2008, 154, 1927–1938. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Campylobacter gracilis and Campylobacter rectus in primary endodontic infections. Int. Endod. J. 2003, 36, 174–180. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum-symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, D.J.; Marsh, P.D.; Watson, G.K.; Allison, C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 1998, 66, 4729–4732. [Google Scholar] [CrossRef] [PubMed]
- Love, R.M. Enterococcus faecalis--a mechanism for its role in endodontic failure. Int. Endod. J. 2001, 34, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Sum, C.; Mohanty, S.; Gupta, P.K.; Kishen, A. Influence of endodontic chemical treatment on Enterococcus faecalis adherence to collagen studied with laser scanning confocal microscopy and optical tweezers: A preliminary study. J. Biomed. Opt. 2008, 13, 044017. [Google Scholar] [CrossRef]
- Cooper, P.R.; Chicca, I.J.; Holder, M.J.; Milward, M.R. Inflammation and Regeneration in the Dentin-pulp Complex: Net Gain or Net Loss? J. Endod. 2017, 43, S87–s94. [Google Scholar] [CrossRef]
- Alhede, M.; Qvortrup, K.; Liebrechts, R.; Hoiby, N.; Givskov, M.; Bjarnsholt, T. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol. Med. Microbiol. 2012, 65, 335–342. [Google Scholar] [CrossRef]
- Schaudinn, C.; Carr, G.; Gorur, A.; Jaramillo, D.; Costerton, J.; Webster, P. Imaging of endodontic biofilms by combined microscopy (FISH/cLSM-SEM). J. Microsc. 2009, 235, 124–127. [Google Scholar] [CrossRef]
- Van Frankenhuyzen, J.K.; Trevors, J.T.; Lee, H.; Flemming, C.A.; Habash, M.B. Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration. J. Microbiol. Methods 2011, 87, 263–272. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Roman, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrodinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Bustin, S.; Nolan, T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Investig. 2017, 47, 756–774. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]






| Organism | Sequence (5′ → 3′) | Reference |
|---|---|---|
| Streptococcus anginosus | fw: ACC AGG TCT TGA CAT CCC GAT GCT A rv: CCA TGC ACC ACC TGTC ACC GA | [31] |
| Streptococcus oralis | fw: ACC AGG TCT TGA CAT CCC TCT GAC C rv: ACCACCTGTCACCTCTGTCCCG | [31] |
| Actinomyces oris | fw: GCC TGT CCC TTT GTG GGT GGG rv: GCG GCT GCT GGC ACG TAG TT | [31] |
| Veillonella dispar | fw: CCC GGG CCT TGT ACA CAC CG rv: CCC ACC GGC TTT GGG CAC TT | [31] |
| Fusobacterium nucleatum | fw: CGC CCG TCA CAC CAC GAG A rv: ACA CCC TCG GAA CAT CCC TCC TTA C | [31] |
| Campylobacter rectus | fw: TCA CCG CCC GTC ACA CCA TG rv: CCG GTT TGG TAT TTG GGC TTC GAG T | [31] |
| Prevotella intermedia | fw: GCG TGC AGA TTG ACG GCC CTA T rv: GGC ACA CGT GCC CGC TTT ACT | [31] |
| Porphyromonas gingivalis | fw: GCG AGA GCC TGA ACC AGC CA rv: ACT CGT ATC GCC CGT TAT TCC CGT A | [31] |
| Treponema denticola | fw: TAA GGG ACA GCT TGC TCA CCC CTA rv: CAC CCA CGC GTT ACT CAC CAG TC | [31] |
| Tannerella forsythia | fw: CGA TGA TAC GCG AGG AAC CTT ACC C rv: CCG AAG GGA AGA AAG CTC TCA CTC T | [31] |
| Enterococcus feacalis | fw: CCG AGT GCT TGC ACT CAA TTG G rv: CTC TTA TGC CAT GCG GCA TAA AC | [34] |
| Probe 1 | Target Organisms | Site 3 | Formamide (%) | Sequence (5′–> 3′) 2 | Reference |
|---|---|---|---|---|---|
| Efae470 | Enterococcus faecalis | 470–489 | 30 | GAT ACC GTC AGG GGA CGT TC | [35] |
| FUS664 | Fusobacterium spp. | 664–683 | 40 | CTT GTA GTT CCG CYT ACC TC | [36] |
| L-Pint649-2 | Prevotella intermedia | 649–667 | 40 | CGT TGC GTG CAC TCA AGT C | [24] |
| Pint649 | Prevotella intermedia | 649–667 | 30–40 | CGT TGC GTG CAC TCA AGT C | [37] |
| Pnig657 | Prevotella nigrescens | 657–675 | 40 | TCC GCC TGC GCT GCG TGT A | [37] |
| Pmic740 | Parvimonas micra | 740–759 | 25 | CTG AGC GTC AGT AAA AGT CC | [38] |
| Sspu439 | Selenomonas sputigena | 439–456 | 40 | CGG TTT TCG TCC CGT GCA | This study |
| TrepG1-679 | Treponemes Cluster 1, (Treponema denticola et rel.) | 679–696 | 40 | GAT TCC ACC CCT ACA CTT | [39] |
| Saur229 | Staphylococcus aureus | 229–246 | 40 | CTA ATG CAG CGC GGA TCC | [40] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukic, D.; Karygianni, L.; Flury, M.; Attin, T.; Thurnheer, T. Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases. Microorganisms 2020, 8, 674. https://doi.org/10.3390/microorganisms8050674
Lukic D, Karygianni L, Flury M, Attin T, Thurnheer T. Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases. Microorganisms. 2020; 8(5):674. https://doi.org/10.3390/microorganisms8050674
Chicago/Turabian StyleLukic, Dejana, Lamprini Karygianni, Manuela Flury, Thomas Attin, and Thomas Thurnheer. 2020. "Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases" Microorganisms 8, no. 5: 674. https://doi.org/10.3390/microorganisms8050674
APA StyleLukic, D., Karygianni, L., Flury, M., Attin, T., & Thurnheer, T. (2020). Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases. Microorganisms, 8(5), 674. https://doi.org/10.3390/microorganisms8050674





