A Plasmid-Encoded FetMP-Fls Iron Uptake System Confers Selective Advantages to Salmonella enterica Serovar Typhimurium in Growth under Iron-Restricted Conditions and for Infection of Mammalian Host Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. In Silico Studies
2.3. Construction of an S. Typhimurium LSP 146/02 fetMP-flsDA Deficient Strain
2.4. Bacterial Growth under Iron-Restricted Conditions
2.5. Intracellular Metal Content Determination by Inductively Coupled Plasma-Mass Spectrometry (ICP-MS)
2.6. Competition Assays
2.7. Infection of Eukaryotic Cell Lines
2.7.1. Invasion of HeLa Cells
2.7.2. Replication within Macrophages
2.8. Complementation Studies and Transformation of S. Typhimurium ATCC 14028 with the Cloned fetMP-flsDA Genes
2.9. Statistical Analysis
3. Results
3.1. Multiple High-Affinity Iron-Uptake Systems are Encoded by the Genome of S. Typhimurium LSP 146/02
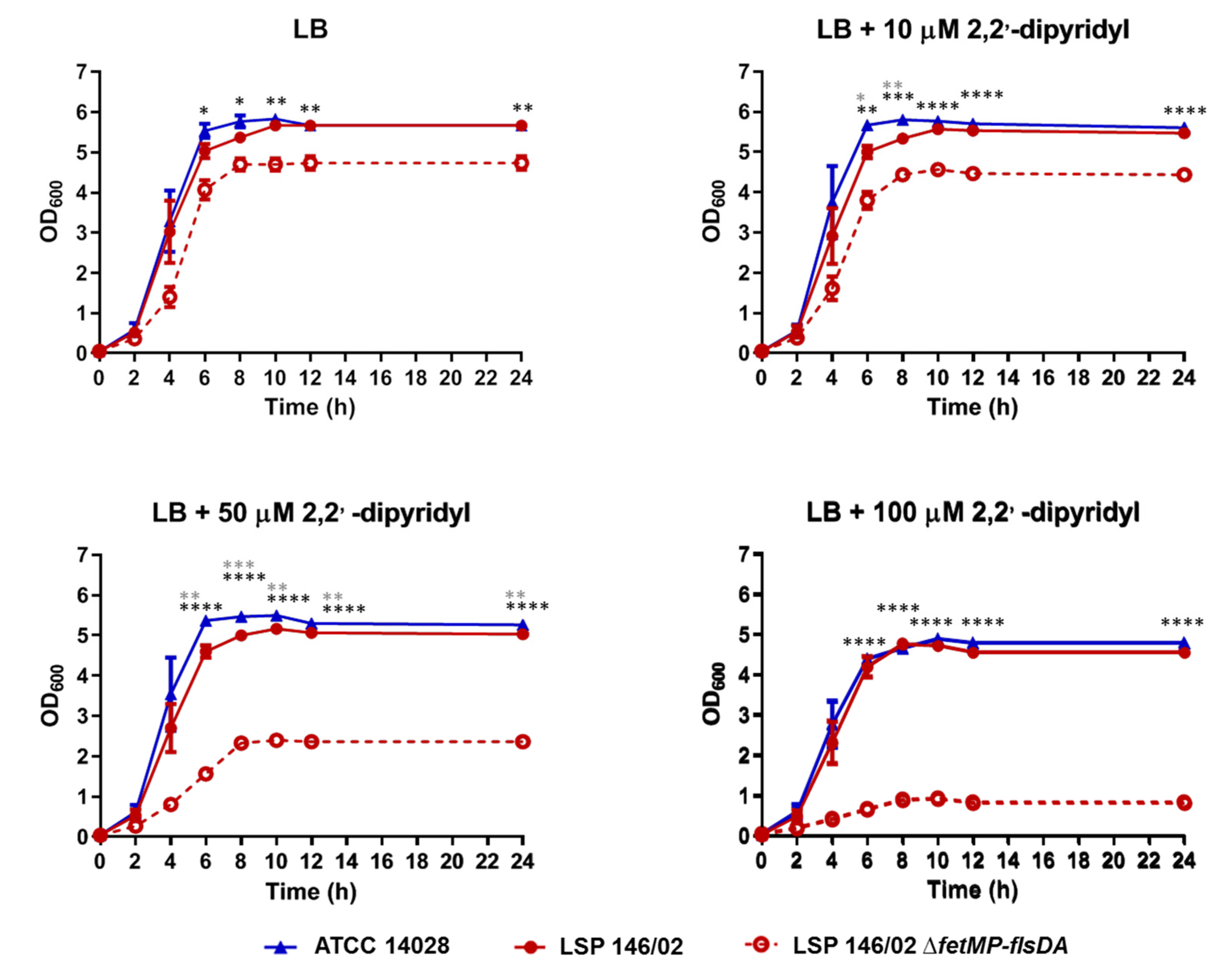
3.2. Growth of the S. Typhimurium LSP 146/02 ΔfetMP-flsDA Strain is Impaired under Iron-Restrictive Conditions
3.3. The Intracellular Iron Content is Higher in S. Typhimurium LSP 146/02 than in the ΔfetMP-flsDA Mutant
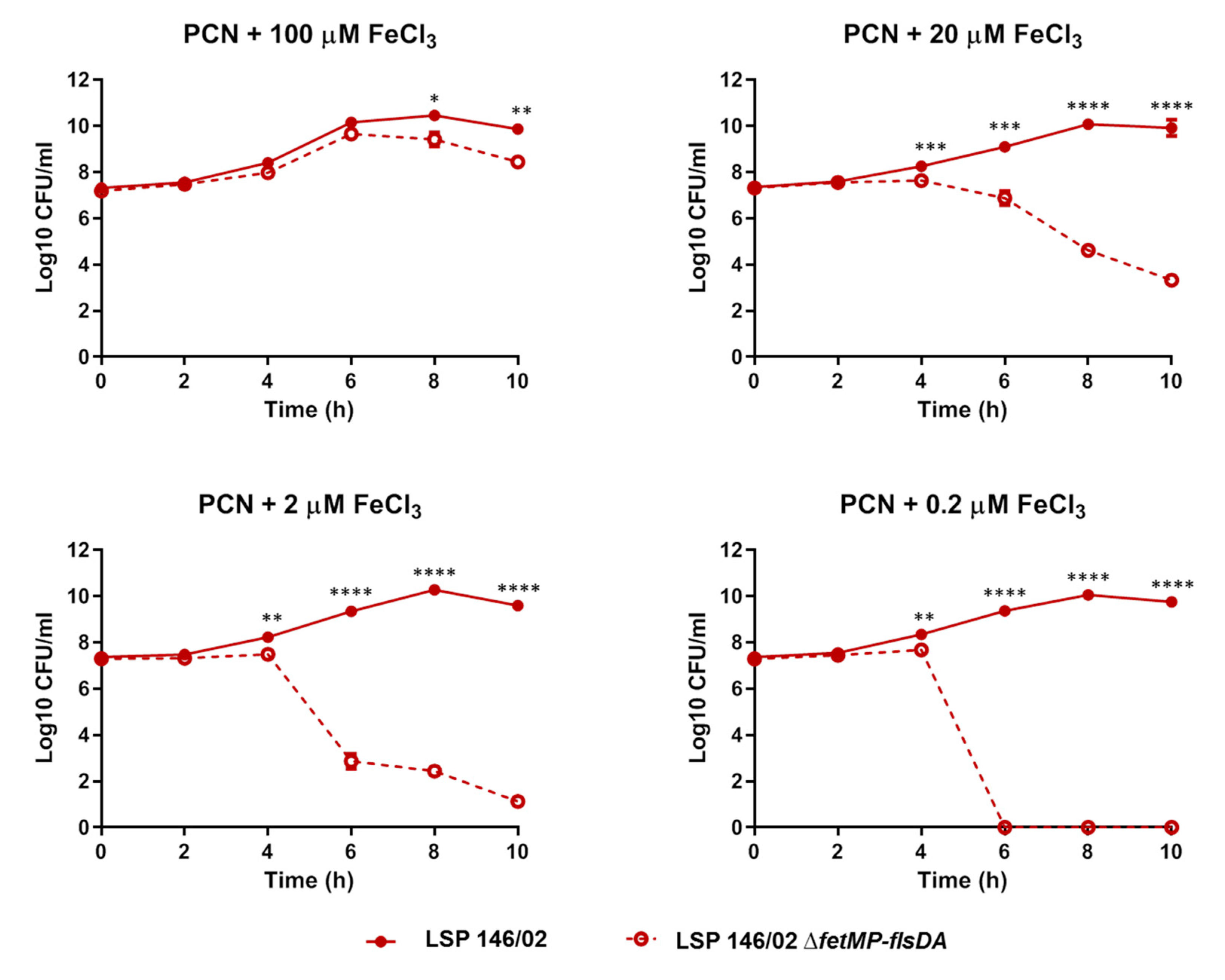
3.4. S. Typhimurium LSP 146/02 Outcompetes its ΔfetMP-flsDA Mutant Strain under Iron Limitation
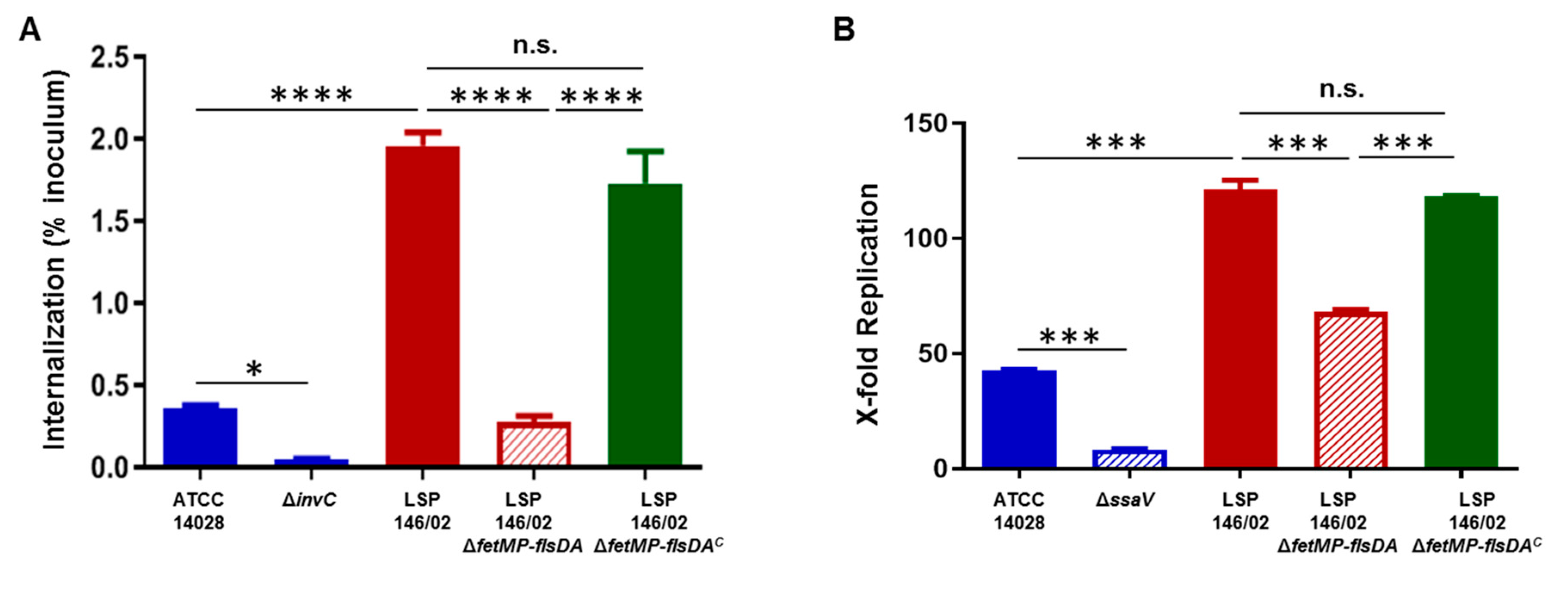
3.5. Invasion of Epithelial Cells and Replication within Macrophages Decrease in the Absence of the fetMP-flsDA Genes
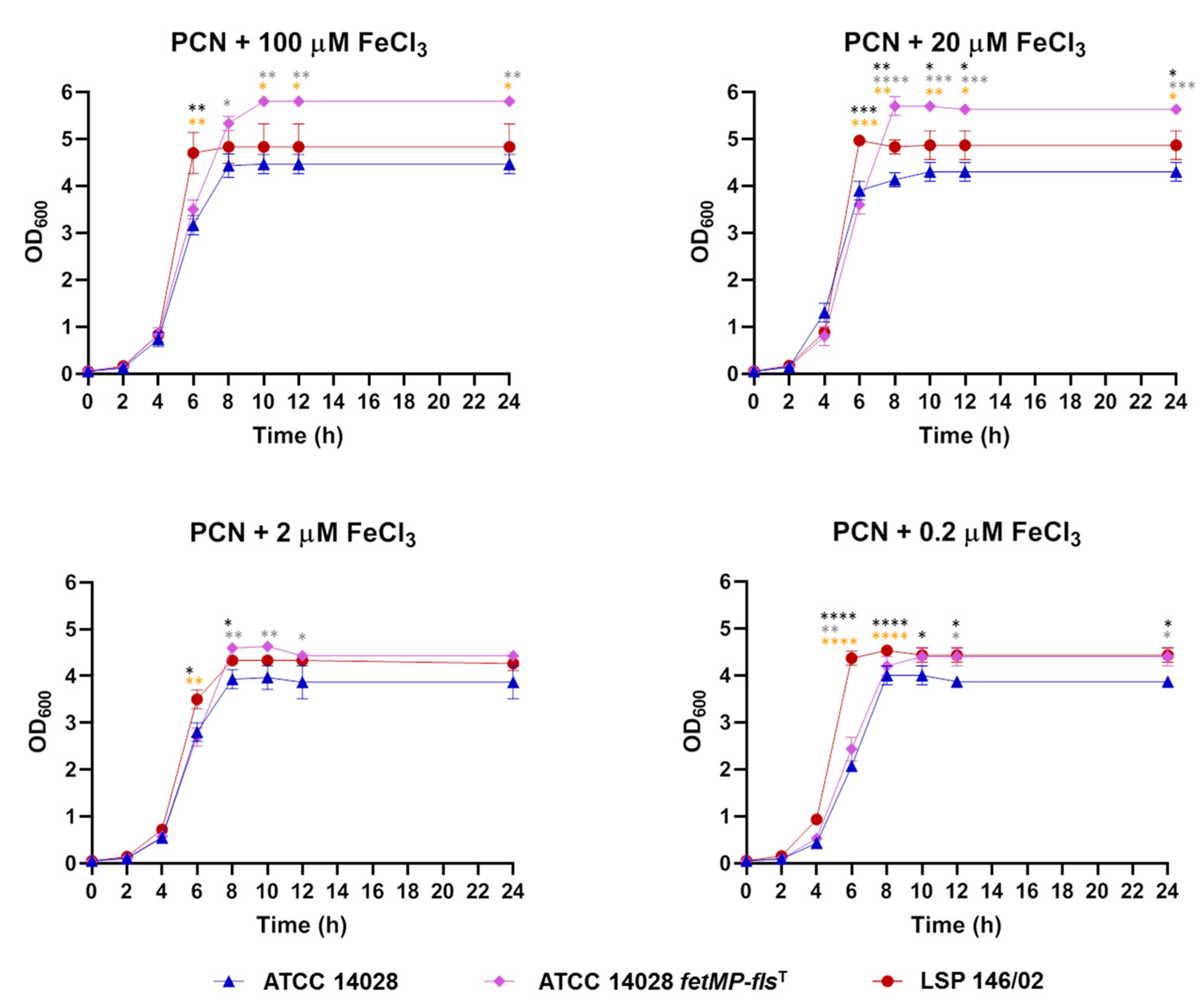
3.6. The FetMP-Fls System of pUO-StVR2 Increases Performance of S. Typhimurium ATCC 14028 under Iron-Limiting Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. International Collaboration on Enteric Disease “Burden of Illness” Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Kortman, G.A.; Roelofs, R.W.; Swinkels, D.W.; de Jonge, M.I.; Burt, S.A.; Tjalsma, H. Iron-induced virulence of Salmonella enterica serovar Typhimurium at the intestinal epithelial interface can be suppressed by carvacrol. Antimicrob. Agents Chemother. 2014, 58, 1664–1670. [Google Scholar] [CrossRef] [PubMed]
- Braun, V. Regulation of iron uptake minimizes iron-mediated oxidative stress. J. Biosci. 1998, 23, 483–489. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.D. Iron and susceptibility to infectious disease. Science 1974, 184, 952–956. [Google Scholar] [CrossRef]
- Kingsley, R.A. Iron Uptake and Metabolism by Salmonella enterica. Ph.D. Thesis, University of Leicester, Leicester, UK, 1997. [Google Scholar]
- Pollack, J.R.; Neilands, J.B. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem. Biophys. Res. Commun. 1970, 38, 989–992. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Tsolis, R.M.; van der Velden, A.W.; Stojiljkovic, I.; Anic, S.; Heffron, F. Identification of a new iron regulated locus of Salmonella typhi. Gene 1996, 183, 207–213. [Google Scholar] [CrossRef]
- Müller, S.I.; Valdebenito, M.; Hantke, K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 2009, 22, 691–695. [Google Scholar] [CrossRef]
- Raffatellu, M.; George, M.D.; Akiyama, Y.; Hornsby, M.J.; Nuccio, S.P.; Paixao, T.A.; Butler, B.P.; Chu, H.; Santos, R.L.; Berger, T.; et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 2009, 5, 476–486. [Google Scholar]
- Kingsley, R.; Rabsch, W.; Stephens, P.; Roberts, M.; Reissbrodt, R.; Williams, P.H. Iron supplying systems of Salmonella in diagnostics, epidemiology and infection. FEMS Immunol. Med. Microbiol. 1995, 11, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Luckey, M.; Pollack, J.R.; Wayne, R.; Ames, B.N.; Neilands, J.B. Iron uptake in Salmonella typhimurium: Utilization of exogenous siderochromes as iron carriers. J. Bacteriol. 1972, 111, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Cartron, M.L.; Maddocks, S.; Gillingham, P.; Craven, C.J.; Andrews, S.C. Feo-transport of ferrous iron into bacteria. Biometals 2006, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Janakiraman, A.; Slauch, J.M. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 2000, 35, 1146–1155. [Google Scholar] [CrossRef]
- Boyer, E.; Bergevin, I.; Malo, D.; Gros, P.; Cellier, M.F. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 2002, 70, 6032–6042. [Google Scholar] [CrossRef]
- Herrero, A.; Rodicio, M.R.; González-Hevia, M.A.; Mendoza, M.C. Molecular epidemiology of emergent multidrug-resistant Salmonella enterica serotype Typhimurium strains carrying the virulence resistance plasmid pUO-StVR2. J. Antimicrob. Chemother. 2006, 57, 39–45. [Google Scholar] [CrossRef]
- Herrero, A.; Rodicio, M.R.; Echeita, M.A.; Mendoza, M.C. Salmonella enterica serotype Typhimurium carrying hybrid virulence-resistance plasmids (pUO-StVR): A new multidrug-resistant group endemic in Spain. Int. J. Med. Microbiol. 2008, 298, 253–261. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination amongst humans and food products of animal origin of a Salmonella typhimurium clone expressing an integron-borne OXA-30 beta-lactamase. J. Antimicrob. Chemother. 2004, 54, 429–434. [Google Scholar] [CrossRef]
- Herrero, A.; Mendoza, M.C.; Threlfall, E.J.; Rodicio, M.R. Detection of Salmonella enterica serovar Typhimurium with pUO-StVR2-like virulence-resistance hybrid plasmids in the United Kingdom. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 1087–1093. [Google Scholar] [CrossRef]
- Beutlich, J.; Rodicio, M.R.; Mendoza, M.C.; García, P.; Kirchner, M.; Luzzi, I.; Mevius, D.; Threlfall, J.; Helmuth, R.; Guerra, B. Med-Vet-Net WP21 Project Group. Salmonella enterica serovar Typhimurium virulence-resistance plasmids derived from the pSLT carrying nonconventional class 1 integrons with dfrA12 gene in their variable region and sul3 in the 3’ conserved segment. Microb. Drug Resist. 2013, 19, 437–445. [Google Scholar] [PubMed]
- Carpenter, C.; Payne, S.M. Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 2014, 133, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Mendoza, M.C.; Rodicio, R.; Rodicio, M.R. Characterization of pUO-StVR2, a virulence-resistance plasmid evolved from the pSLT virulence plasmid of Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 2008, 52, 4514–4517. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, H.; Sekizuka, T.; Nakaya, H.; Taguchi, M.; Oguchi, A.; Ichikawa, N.; Nishiko, R.; Yamazaki, S.; Fujita, N.; Watanabe, H.; et al. Whole-genome analysis of Salmonella enterica serovar Typhimurium T000240 reveals the acquisition of a genomic island involved in multidrug resistance via IS1 derivatives on the chromosome. Antimicrob. Agents Chemother. 2011, 55, 623–630. [Google Scholar] [CrossRef]
- Liu, M.M.; Boinett, C.J.; Chan, A.C.K.; Parkhill, J.; Murphy, M.E.P.; Gaynor, E.C. Investigating the Campylobacter jejuni transcriptional response to host intestinal extracts reveals the involvement of a widely conserved iron uptake system. MBio 2018, 9, e01347-18. [Google Scholar] [CrossRef]
- Koch, D.; Chan, A.C.; Murphy, M.E.; Lilie, H.; Grass, G.; Nies, D.H. Characterization of a dipartite iron uptake system from uropathogenic Escherichia coli strain F11. J. Biol. Chem. 2011, 286, 25317–25330. [Google Scholar] [CrossRef]
- Chan, A.C.; Doukov, T.I.; Scofield, M.; Tom-Yew, S.A.; Ramin, A.B.; Mackichan, J.K.; Gaynor, E.C.; Murphy, M.E. Structure and function of P19, a high-affinity iron transporter of the human pathogen Campylobacter jejuni. J. Mol. Biol. 2010, 401, 590–604. [Google Scholar] [CrossRef]
- Fetherston, J.D.; Mier, I.; Truszczynska, H.; Perry, R.D. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun. 2012, 80, 3880–3891. [Google Scholar] [CrossRef]
- Gerlach, R.G.; Cláudio, N.; Rohde, M.; Jäckel, D.; Wagner, C.; Hensel, M. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell. Microbiol. 2008, 10, 2364–2376. [Google Scholar] [CrossRef]
- Shea, J.E.; Hensel, M.; Gleeson, C.; Holden, D.W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1996, 93, 2593–2597. [Google Scholar] [CrossRef]
- Taylor, R.G.; Walker, D.C.; McInnes, R.R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993, 21, 1677–1678. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Douard, G.; Targant, H.; Meunier, D.; Madec, J.Y.; Cloeckaert, A. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 2008, 75, 359–361. [Google Scholar] [CrossRef]
- Deiwick, J.; Nikolaus, T.; Erdogan, S.; Hensel, M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 1999, 31, 1759–1773. [Google Scholar] [CrossRef]
- Hassan, K.A.; Pederick, V.G.; Elbourne, L.D.; Paulsen, I.T.; Paton, J.C.; McDevitt, C.A.; Eijkelkamp, B.A. Zinc stress induces copper depletion in Acinetobacter baumannii. BMC Microbiol. 2017, 17, 59. [Google Scholar] [CrossRef]
- Kasahara, M.; Nakata, A.; Shinagawa, H. Molecular analysis of the Salmonella typhimurium phoN gene, which encodes nonspecific acid phosphatase. J. Bacteriol. 1991, 173, 6760–6765. [Google Scholar] [CrossRef]
- García, P.; Guerra, B.; Bances, M.; Mendoza, M.C.; Rodicio, M.R. IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype 4,[5],12:i. J. Antimicrob. Chemother. 2011, 66, 543–549. [Google Scholar] [CrossRef]
- Wang, R.F.; Kushner, S.R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 1991, 100, 195–199. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 2001. [Google Scholar]
- Porcheron, G.; Garénaux, A.; Proulx, J.; Sabri, M.; Dozois, C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: Correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 2013, 3, 90. [Google Scholar] [CrossRef]
- Dichtl, S.; Demetz, E.; Haschka, D.; Tymoszuk, P.; Petzer, V.; Nairz, M.; Seifert, M.; Hoffmann, A.; Brigo, N.; Würzner, R.; et al. Dopamine is a siderophore-like iron chelator that promotes Salmonella enterica serovar Typhimurium virulence in mice. mBio 2019, 10, e02624-18. [Google Scholar] [CrossRef] [PubMed]
- Hurley, D.; McCusker, M.P.; Fanning, S.; Martins, M. Salmonella-host interactions—Modulation of the host innate immune system. Front. Immunol. 2014, 5, 481. [Google Scholar] [PubMed]



| Strain | Fe [µg/g dw] | Mn [µg/g dw] | Zn [µg/g dw] |
|---|---|---|---|
| LSP 146/02 | 1516.48 ± 184.32 | 39.58 ± 9.96 | 60.05 ± 3.69 |
| LSP 146/02 ΔfetMP-flsDA | 280.69 ± 90.68 | 31.78 ± 3.89 | 60.22 ± 7.72 |
| p-value | ˂ 0.0005 | 0.2747 | 0.9742 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García, V.; Herrero-Fresno, A.; Rodicio, R.; Felipe-López, A.; Montero, I.; Olsen, J.E.; Hensel, M.; Rodicio, M.R. A Plasmid-Encoded FetMP-Fls Iron Uptake System Confers Selective Advantages to Salmonella enterica Serovar Typhimurium in Growth under Iron-Restricted Conditions and for Infection of Mammalian Host Cells. Microorganisms 2020, 8, 630. https://doi.org/10.3390/microorganisms8050630
García V, Herrero-Fresno A, Rodicio R, Felipe-López A, Montero I, Olsen JE, Hensel M, Rodicio MR. A Plasmid-Encoded FetMP-Fls Iron Uptake System Confers Selective Advantages to Salmonella enterica Serovar Typhimurium in Growth under Iron-Restricted Conditions and for Infection of Mammalian Host Cells. Microorganisms. 2020; 8(5):630. https://doi.org/10.3390/microorganisms8050630
Chicago/Turabian StyleGarcía, Vanesa, Ana Herrero-Fresno, Rosaura Rodicio, Alfonso Felipe-López, Ignacio Montero, John E. Olsen, Michael Hensel, and María Rosario Rodicio. 2020. "A Plasmid-Encoded FetMP-Fls Iron Uptake System Confers Selective Advantages to Salmonella enterica Serovar Typhimurium in Growth under Iron-Restricted Conditions and for Infection of Mammalian Host Cells" Microorganisms 8, no. 5: 630. https://doi.org/10.3390/microorganisms8050630
APA StyleGarcía, V., Herrero-Fresno, A., Rodicio, R., Felipe-López, A., Montero, I., Olsen, J. E., Hensel, M., & Rodicio, M. R. (2020). A Plasmid-Encoded FetMP-Fls Iron Uptake System Confers Selective Advantages to Salmonella enterica Serovar Typhimurium in Growth under Iron-Restricted Conditions and for Infection of Mammalian Host Cells. Microorganisms, 8(5), 630. https://doi.org/10.3390/microorganisms8050630







