Improved Protocol for DNA Extraction from Subsoils Using Phosphate Lysis Buffer
Abstract
1. Introduction
2. Materials and Methods
2.1. Optimization of DNA Extraction from Subsoil
2.1.1. Subsoil Collection for Optimization
2.1.2. Subsoil DNA Extraction Using CTAB Buffer
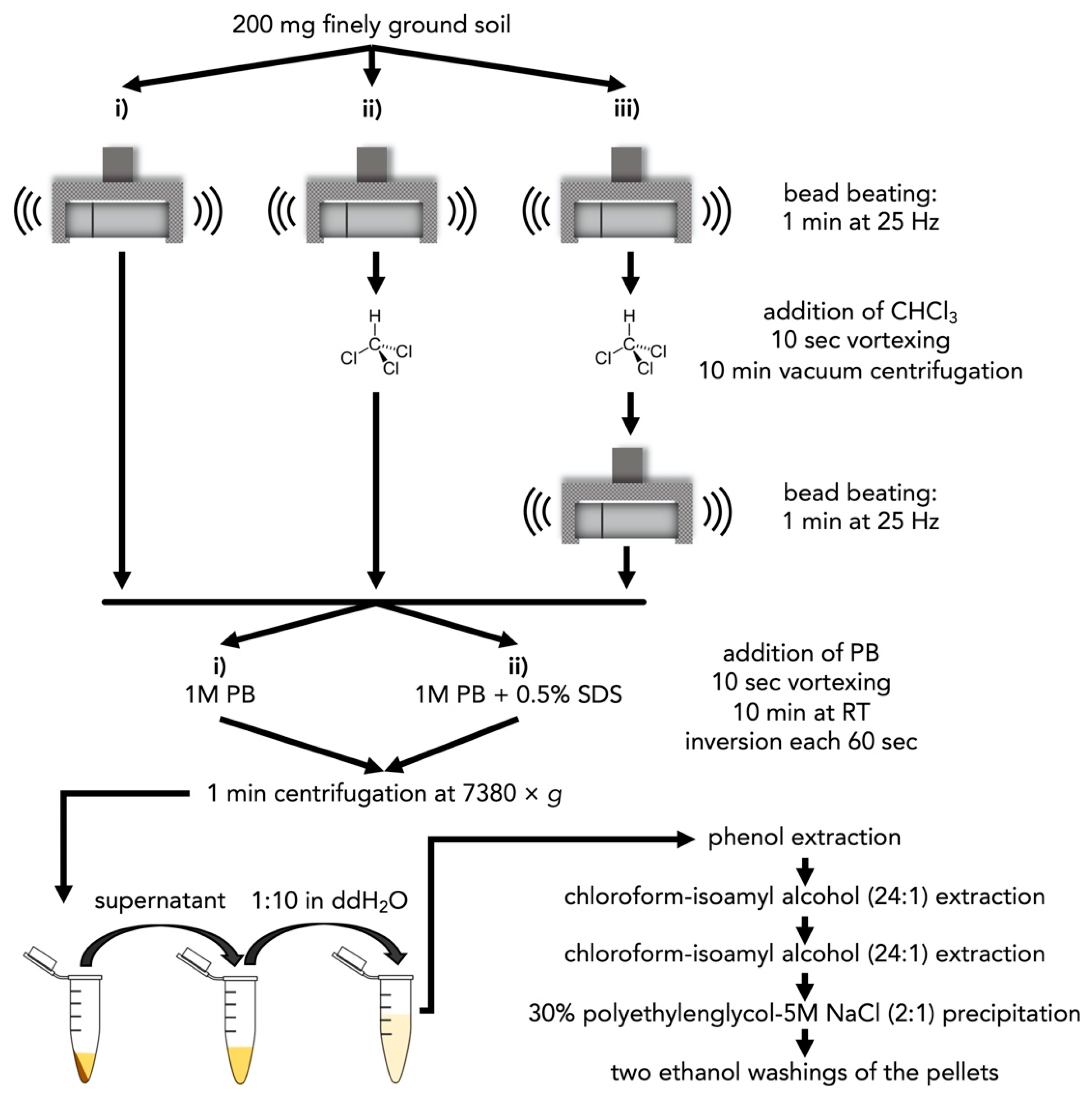
2.1.3. Subsoil DNA Extraction Using Phosphate Buffer
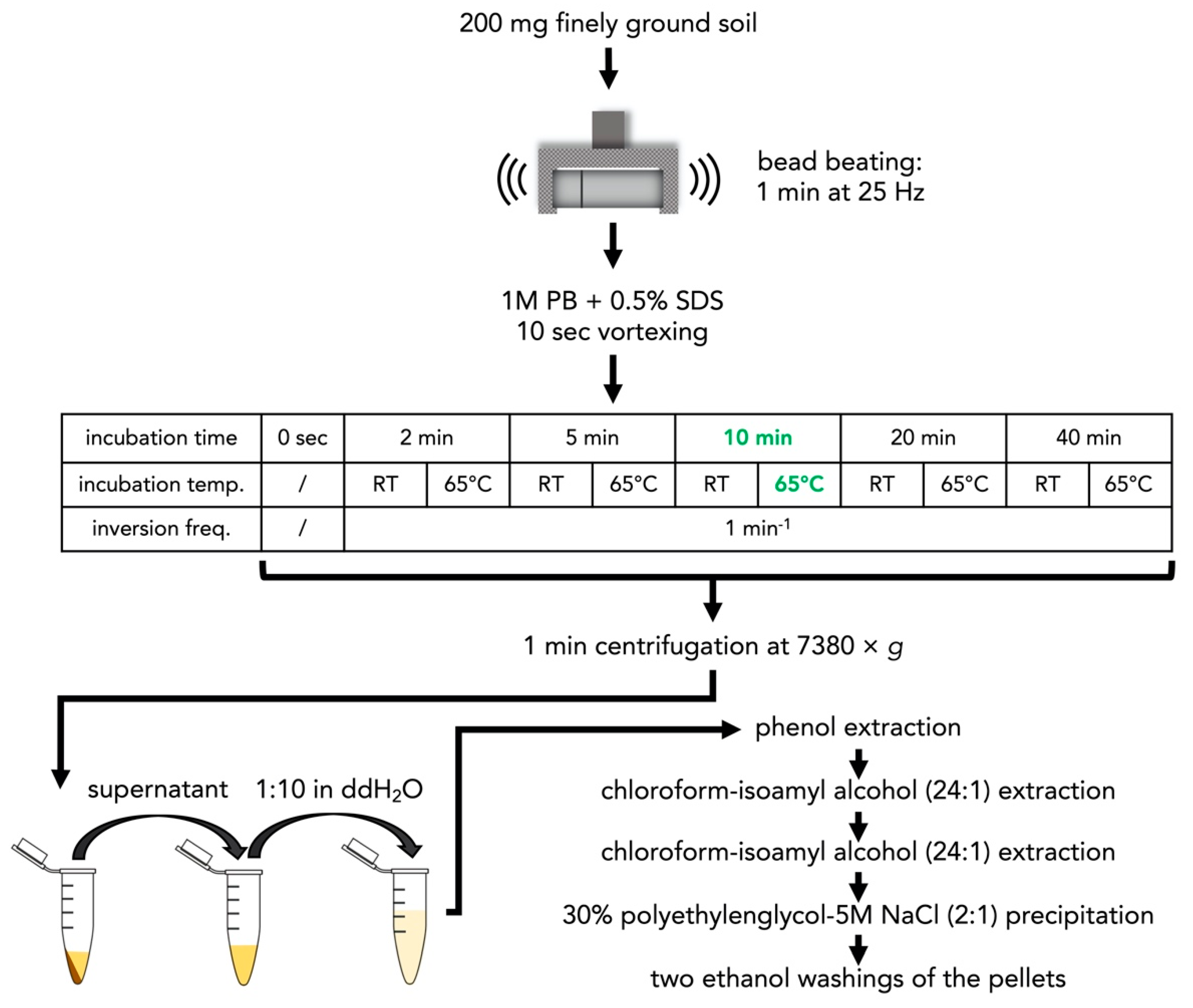
2.1.4. Optimization of the Incubation Temperature and Time in the Phosphate Buffer
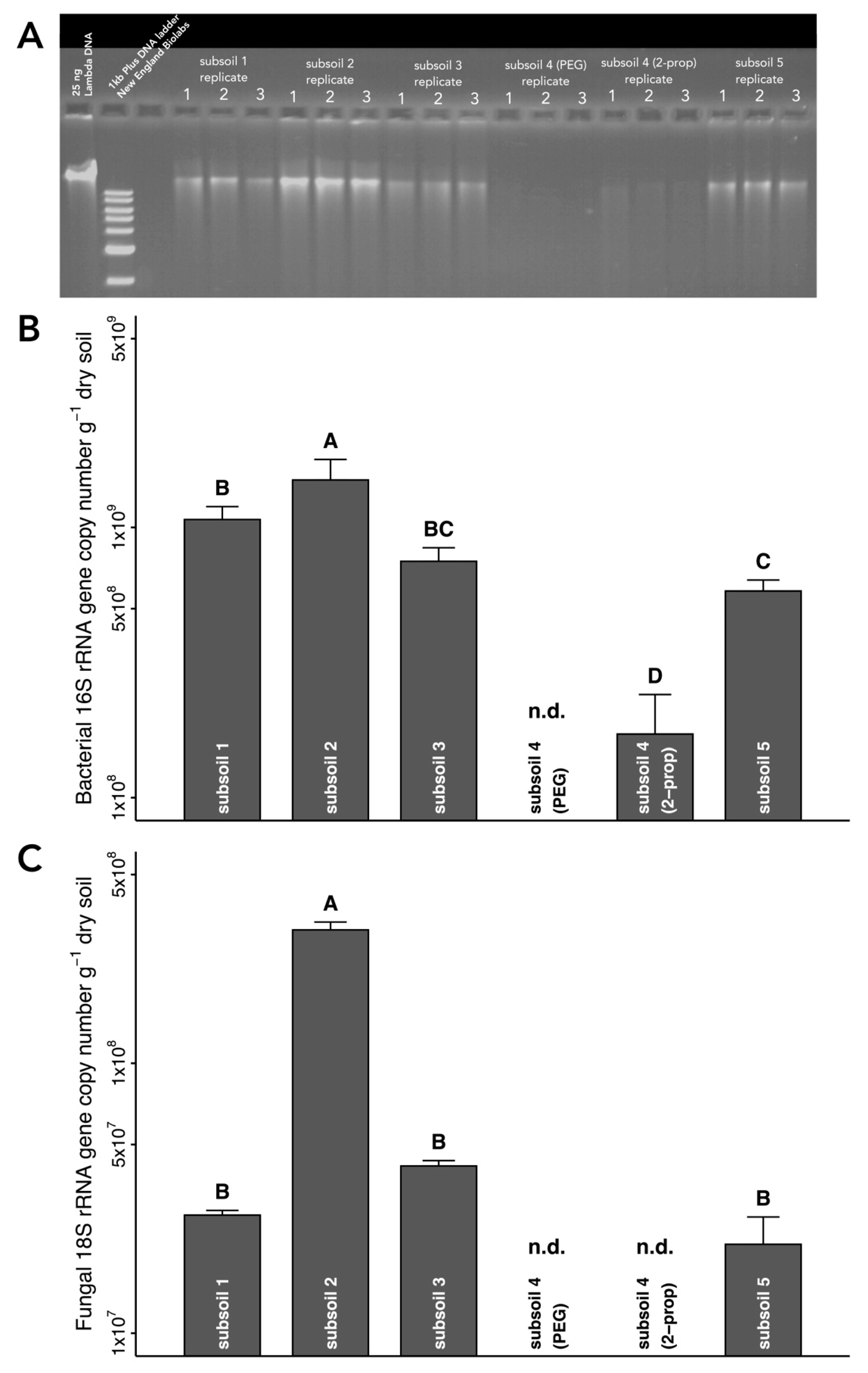
2.2. DNA Extraction from Different Types of Subsoil
2.3. Quantification of Soil Bacteria and Fungi
2.4. DNA Amplification Inhibition Test
2.5. Interlaboratory Comparison
2.6. Statistical Analysis
3. Results
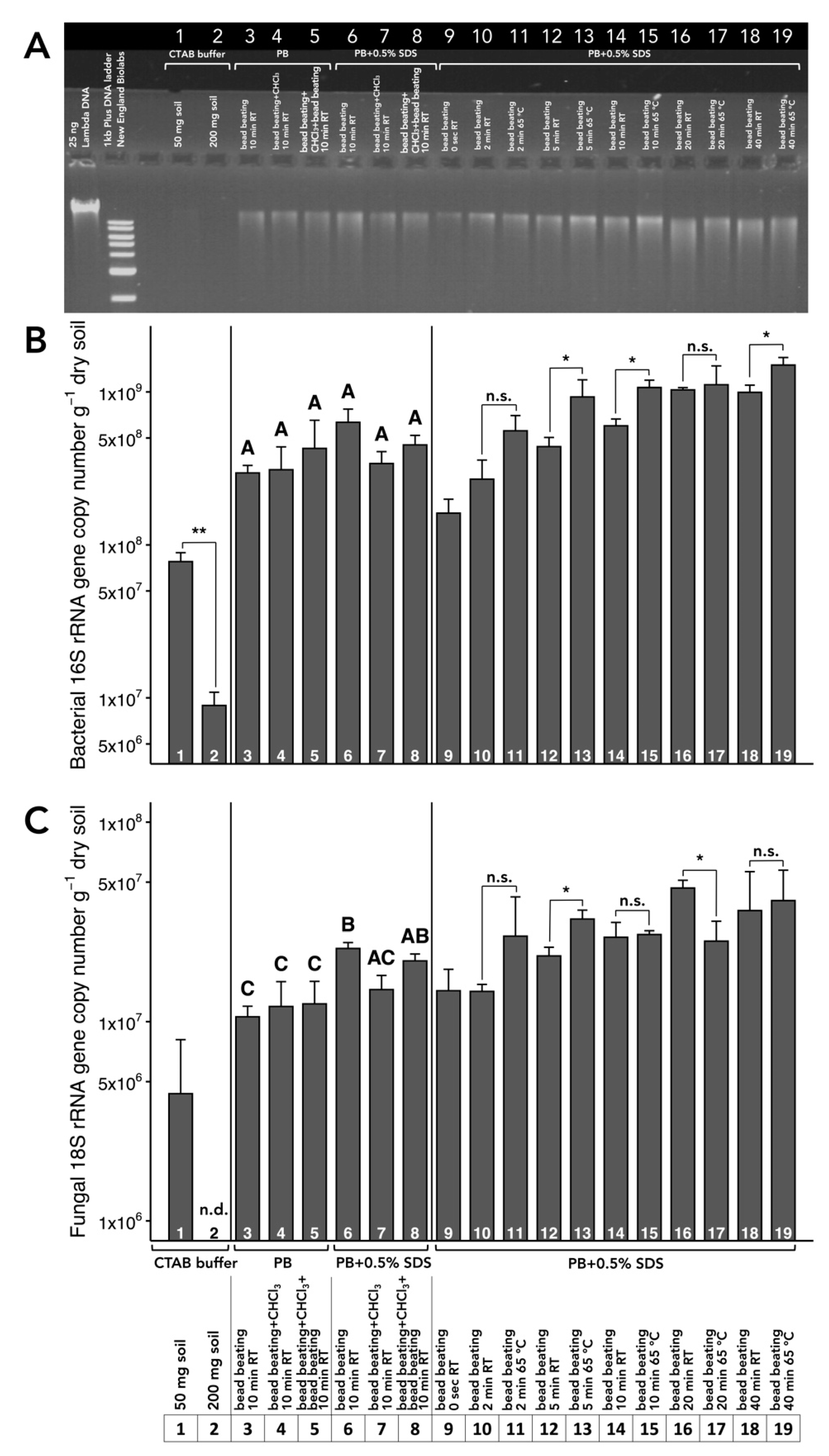
3.1. Choice of Extraction Buffer
3.2. Optimization of Incubation Temperature and Time
3.3. DNA Extraction from Different Subsoils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tebrügge, F.; Düring, R.-A. Reducing tillage intensity—A review of results from a long-term study in Germany. Soil Tillage Res. 1999, 53, 15–28. [Google Scholar] [CrossRef]
- Sosa-Hernández, M.A.; Leifheit, E.F.; Ingraffia, R.; Rillig, M.C. Subsoil Arbuscular Mycorrhizal Fungi for Sustainability and Climate-Smart Agriculture: A Solution Right under Our Feet? Front. Microbiol. 2019, 10, 744. [Google Scholar] [CrossRef] [PubMed]
- Frelih-Larsen, A.; Hinzmann, M.; Ittner, S. The ‘Invisible’ Subsoil: An Exploratory View of Societal Acceptance of Subsoil Management in Germany. Sustainability 2018, 10, 3006. [Google Scholar] [CrossRef]
- Holden, P.A.; Fierer, N. Microbial Processes in the Vadose Zone. Vadose Zone J. 2005, 4, 1–21. [Google Scholar] [CrossRef]
- von Lützow, M.; Kögel-Knabner, I.; Ekschmitt, K.; Matzner, E.; Guggenberger, G.; Marschner, B.; Flessa, H. Stabilization of organic matter in temperate soils: Mechanisms and their relevance under different soil conditions—A review. Eur. J. Soil Sci. 2006, 57, 426–445. [Google Scholar] [CrossRef]
- Fontaine, S.; Barot, S.; Barré, P.; Bdioui, N.; Mary, B.; Rumpel, C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 2007, 450, 277–280. [Google Scholar] [CrossRef]
- Xiang, S.-R.; Doyle, A.; Holden, P.A.; Schimel, J.P. Drying and rewetting effects on C and N mineralization and microbial activity in surface and subsurface California grassland soils. Soil Biol. Biochem. 2008, 40, 2281–2289. [Google Scholar] [CrossRef]
- Chabbi, A.; Kögel-Knabner, I.; Rumpel, C. Stabilised carbon in subsoil horizons is located in spatially distinct parts of the soil profile. Soil Biol. Biochem. 2009, 41, 256–261. [Google Scholar] [CrossRef]
- Fierer, N.; Allen, A.S.; Schimel, J.P.; Holden, P.A. Controls on microbial CO2 production: A comparison of surface and subsurface soil horizons. Glob. Chang. Biol. 2003, 9, 1322–1332. [Google Scholar] [CrossRef]
- Oehl, F.; Sieverding, E.; Ineichen, K.; Ris, E.A.; Boller, T.; Wiemken, A. Community structure of arbuscular mycorrhizal fungi at different soil depths in extensively and intensively managed agroecosystems. New Phytol. 2005, 165, 273–283. [Google Scholar] [CrossRef]
- Sagova-Mareckova, M.; Zadorova, T.; Penizek, V.; Omelka, M.; Tejnecky, V.; Pruchova, P.; Chuman, T.; Drabek, O.; Buresova, A.; Vanek, A.; et al. The structure of bacterial communities along two vertical profiles of a deep colluvial soil. Soil Biol. Biochem. 2016, 101, 65–73. [Google Scholar] [CrossRef]
- Kautz, T.; Perkons, U.; Athmann, M.; Pude, R.; Köpke, U. Barley roots are not constrained to large-sized biopores in the subsoil of a deep Haplic Luvisol. Biol. Fertil. Soils 2013, 49, 959–963. [Google Scholar] [CrossRef]
- Hobley, E.; Bauke, S.; Steffens, M.; Amelung, W.; Kögel-Knabner, I. Hyperspectral Imaging of Soil Cores Reveals Greatest C Storage in Subsoil Biopores; Geophysical Research Abstracts; European Geosciences Union: Strasbourg, France, 2018; Volume 20, p. 12765. [Google Scholar]
- Uksa, M.; Schloter, M.; Endesfelder, D.; Kublik, S.; Engel, M.; Kautz, T.; Köpke, U.; Fischer, D. Prokaryotes in Subsoil—Evidence for a Strong Spatial Separation of Different Phyla by Analysing Co-occurrence Networks. Front. Microbiol. 2015, 6, 1269. [Google Scholar] [CrossRef] [PubMed]
- Lavahun, M.F.E.; Joergensen, R.G.; Meyer, B. Activity and biomass of soil microorganisms at different depths. Biol. Fertil. Soils 1996, 23, 38–42. [Google Scholar] [CrossRef]
- Taylor, J.P.; Wilson, B.; Mills, M.S.; Burns, R.G. Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol. Biochem. 2002, 34, 387–401. [Google Scholar] [CrossRef]
- Zhang, B.; Penton, C.R.; Xue, C.; Quensen, J.F.; Roley, S.S.; Guo, J.; Garoutte, A.; Zheng, T.; Tiedje, J.M. Soil depth and crop determinants of bacterial communities under ten biofuel cropping systems. Soil Biol. Biochem. 2017, 112, 140–152. [Google Scholar] [CrossRef]
- Sosa-Hernández, M.A.; Roy, J.; Hempel, S.; Kautz, T.; Köpke, U.; Uksa, M.; Schloter, M.; Caruso, T.; Rillig, M.C. Subsoil arbuscular mycorrhizal fungal communities in arable soil differ from those in topsoil. Soil Biol. Biochem. 2018, 117, 83–86. [Google Scholar] [CrossRef]
- Tebbe, C.C.; Vahjen, W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 1993, 59, 2657–2665. [Google Scholar] [CrossRef]
- Ranjard, L.; Poly, F.; Combrisson, J.; Richaume, A.; Nazaret, S. A single procedure to recover DNA from the surface or inside aggregates and in various size fractions of soil suitable for PCR-based assays of bacterial communities. Eur. J. Soil Biol. 1998, 34, 89–97. [Google Scholar] [CrossRef]
- Frostegård, Å.; Courtois, S.; Ramisse, V.; Clerc, S.; Bernillon, D.; Le Gall, F.; Jeannin, P.; Nesme, X.; Simonet, P. Quantification of Bias Related to the Extraction of DNA Directly from Soils. Appl. Environ. Microbiol. 1999, 65, 5409–5420. [Google Scholar] [CrossRef]
- Hurt, R.A.; Robeson, M.S.; Shakya, M.; Moberly, J.G.; Vishnivetskaya, T.A.; Gu, B.; Elias, D.A. Improved yield of high molecular weight DNA coincides with increased microbial diversity access from iron oxide cemented sub-surface clay environments. PLoS ONE 2014, 9, e102826. [Google Scholar] [CrossRef] [PubMed]
- Goring, C.A.I.; Bartholomew, W.V. Adsorption of Mononucleotides, Nucleic Acids, and Nucleoproteins by Clays. Soil Sci. 1952, 74, 149. [Google Scholar] [CrossRef]
- Greaves, M.P.; Wilson, M.J. The adsorption of nucleic acids by montmorillonite. Soil Biol. Biochem. 1969, 1, 317–323. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Gustin, D.; Lewis, R.J. DNA adsorption to soils and sediments. Environ. Sci. Technol. 1988, 22, 982. [Google Scholar] [CrossRef]
- Khanna, M.; Stotzky, G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 1992, 58, 1930–1939. [Google Scholar] [CrossRef]
- Ogram, A.V.; Mathot, M.L.; Harsh, J.B.; Boyle, J.; Pettigrew, C.A. Effects of DNA Polymer Length on Its Adsorption to Soils. Appl. Environ. Microbiol. 1994, 60, 393–396. [Google Scholar] [CrossRef]
- Pietramellara, G.; Dal Canto, L.; Vettori, C.; Gallori, E.; Nannipieri, P. Effects of air-drying and wetting cycles on the transforming ability of DNA bound on clay minerals. Soil Biol. Biochem. 1997, 29, 55–61. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Layton, A.C.; Lau, M.C.Y.; Chauhan, A.; Cheng, K.R.; Meyers, A.J.; Murphy, J.R.; Rogers, A.W.; Saarunya, G.S.; Williams, D.E.; et al. Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS Microbiol. Ecol. 2014, 87, 217–230. [Google Scholar] [CrossRef]
- Waring, B.G.; Álvarez-Cansino, L.; Barry, K.E.; Becklund, K.K.; Dale, S.; Gei, M.G.; Keller, A.B.; Lopez, O.R.; Markesteijn, L.; Mangan, S.; et al. Pervasive and strong effects of plants on soil chemistry: A meta-analysis of individual plant ‘Zinke’ effects. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151001. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef]
- Hurt, R.A.; Qiu, X.; Wu, L.; Roh, Y.; Palumbo, A.V.; Tiedje, J.M.; Zhou, J. Simultaneous Recovery of RNA and DNA from Soils and Sediments. Appl. Environ. Microbiol. 2001, 67, 4495–4503. [Google Scholar] [CrossRef] [PubMed]
- Sagova-Mareckova, M.; Cermak, L.; Novotna, J.; Plhackova, K.; Forstova, J.; Kopecky, J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl. Environ. Microbiol. 2008, 74, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xu, Z.; Hughes, J. Pre-lysis washing improves DNA extraction from a forest soil. Soil Biol. Biochem. 2005, 37, 2337–2341. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Barkay, T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Cai, P.; Huang, Q.; Zhang, X.; Chen, H. Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol. Soil Biol. Biochem. 2006, 38, 471–476. [Google Scholar] [CrossRef]
- Brandfass, C.; Karlovsky, P. Upscaled CTAB-Based DNA Extraction and Real-Time PCR Assays for Fusarium culmorum and F. graminearum DNA in Plant Material with Reduced Sampling Error. Int. J. Mol. Sci. 2008, 9, 2306–2321. [Google Scholar] [CrossRef]
- Chourey, K.; Jansson, J.; VerBerkmoes, N.; Shah, M.; Chavarria, K.L.; Tom, L.M.; Brodie, E.L.; Hettich, R.L. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J. Proteome Res. 2010, 9, 6615–6622. [Google Scholar] [CrossRef]
- Chatterjee, A.; Moulik, S.P.; Majhi, P.R.; Sanyal, S.K. Studies on surfactant–biopolymer interaction. I. Microcalorimetric investigation on the interaction of cetyltrimethylammonium bromide (CTAB) and sodium dodecylsulfate (SDS) with gelatin (Gn), lysozyme (Lz) and deoxyribonucleic acid (DNA). Biophys. Chem. 2002, 98, 313–327. [Google Scholar] [CrossRef]
- Beule, L.; Corre, M.D.; Schmidt, M.; Göbel, L.; Veldkamp, E.; Karlovsky, P. Conversion of monoculture cropland and open grassland to agroforestry alters the abundance of soil bacteria, fungi and soil-N-cycling genes. PLoS ONE 2019, 14, e0218779. [Google Scholar] [CrossRef]
- Beule, L.; Lehtsaar, E.; Corre, M.D.; Schmidt, M.; Veldkamp, E.; Karlovsky, P. Poplar Rows in Temperate Agroforestry Croplands Promote Bacteria, Fungi, and Denitrification Genes in Soils. Front. Microbiol. 2020, 10, 3108. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Vainio, E.J.; Hantula, J. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 2000, 104, 927–936. [Google Scholar] [CrossRef]
- Eynck, C.; Koopmann, B.; Grunewaldt-Stoecker, G.; Karlovsky, P.; von Tiedemann, A. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 2007, 118, 259–274. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.r-project.org/ (accessed on 12 March 2020).
- Hou, Y.; Wu, P.; Huang, Z.; Ruan, B.; Liu, P.; Zhu, N. Successful intercalation of DNA into CTAB-modified clay minerals for gene protection. J. Mater. Sci. 2014, 49, 7273–7281. [Google Scholar] [CrossRef]
- Emmons, A.L.; DeBruyn, J.M.; Mundorff, A.Z.; Cobaugh, K.L.; Cabana, G.S. The persistence of human DNA in soil following surface decomposition. Sci. Justice 2017, 57, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Choi, J.-H.; Son, A. Necessity of purification during bacterial DNA extraction with environmental soils. Environ. Health Toxicol. 2017, 32, e2017013. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, M.R.; Veraart, A.J.; de Hollander, M.; Smidt, H.; van Veen, J.A.; Kuramae, E.E. Successive DNA extractions improve characterization of soil microbial communities. PeerJ 2017, 5, e2915. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, K.; Takeuchi, M.; Arima, H.; Tsujiguchi, T. Development of a method to extract protozoan DNA from black soil. Parasite Epidemiol. Control 2019, 4, e00081. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Murat, C.; Deveau, A.; Tacon, F.L.; Frey-Klett, P.; Uroz, S. An improved method compatible with metagenomic analyses to extract genomic DNA from soils in Tuber melanosporum orchards. J. Appl. Microbiol. 2013, 115, 163–170. [Google Scholar] [CrossRef]
- Braid, M.D.; Daniels, L.M.; Kitts, C.L. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 2003, 52, 389–393. [Google Scholar] [CrossRef]
- Van den Boogert, P.H.J.F.; van Gent-Pelzer, M.P.E.; Bonants, P.J.M.; De Boer, S.H.; Wander, J.G.N.; Lévesque, C.A.; van Leeuwen, G.C.M.; Baayen, R.P. Development of PCR-based Detection Methods for the Quarantine Phytopathogen Synchytrium endobioticum, Causal Agent of Potato Wart Disease. Eur. J. Plant Pathol. 2005, 113, 47–57. [Google Scholar] [CrossRef]
- Van Gent-Pelzer, M.P.E.; Krijger, M.; Bonants, P.J.M. Improved real-time PCR assay for detection of the quarantine potato pathogen, Synchytrium endobioticum, in zonal centrifuge extracts from soil and in plants. Eur. J. Plant Pathol. 2010, 126, 129. [Google Scholar] [CrossRef]
- Gonzalez-Franco, A.C.; Robles-Hernandez, L.; Nuñez-Barrios, A.; Strap, J.L.; Crawford, D.L. Molecular and cultural analysis of seasonal actinomycetes in soils from Artemisia tridentata habitat. Phyton (Buenos Aires) 2009, 78, 83–90. [Google Scholar]
- Arbeli, Z.; Fuentes, C.L. Improved purification and PCR amplification of DNA from environmental samples. FEMS Microbiol. Lett. 2007, 272, 269–275. [Google Scholar] [CrossRef]
- Purdy, K.J.; Embley, T.M.; Takii, S.; Nedwell, D.B. Rapid Extraction of DNA and rRNA from Sediments by a Novel Hydroxyapatite Spin-Column Method. Appl. Environ. Microbiol. 1996, 62, 3905–3907. [Google Scholar] [CrossRef]
- LaMontagne, M.G.; Michel, F.C.; Holden, P.A.; Reddy, C.A. Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J. Microbiol. Methods 2002, 49, 255–264. [Google Scholar] [CrossRef]
- Yeates, C.; Gillings, M.R.; Davison, A.D.; Altavilla, N.; Veal, D.A. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proced. Online 1998, 1, 40–47. [Google Scholar] [CrossRef]
- Cullen, D.W.; Hirsch, P.R. Simple and rapid method fordirect extraction of microbial DNA fromsoil for PCR. Soil Biol. Biochem. 1998, 30, 983–993. [Google Scholar] [CrossRef]
- Dunn, I.S.; Blattner, F.R. Charons 36 to 40: Multi enzyme, high capacity, recombination deficient replacement vectors with polylinkers and polystuffers. Nucleic Acids Res. 1987, 15, 2677–2698. [Google Scholar] [CrossRef]
- Paithankar, K.R.; Prasad, K.S. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 1991, 19, 1346. [Google Scholar] [CrossRef]
- Gaillard, C.; Strauss, F. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990, 18, 378. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; van der Eb, A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973, 52, 456–467. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Precipitation of DNA with Ethanol. Cold Spring Harb. Protoc. 2016. [Google Scholar] [CrossRef] [PubMed]
- Woldringh, C.L. Lysis of the cell membrane of Escherichia coli K12 by ionic detergents. Biochim. Biophys. Acta (BBA)—Nucleic Acids Protein Synth. 1970, 224, 288–290. [Google Scholar] [CrossRef]
- Woldringh, C.L.; Van Iterson, W. Effects of Treatment with Sodium Dodecyl Sulfate on the Ultrastructure of Escherichia coli. J. Bacteriol. 1972, 111, 801–813. [Google Scholar] [CrossRef]
- Kabir, S.; Rajendran, N.; Amemiya, T.; Itoh, K. Real-time quantitative PCR assay on bacterial DNA: In a model soil system and environmental samples. J. Gen. Appl. Microbiol. 2003, 49, 101–109. [Google Scholar] [CrossRef]
- Miller, D.N.; Bryant, J.E.; Madsen, E.L.; Ghiorse, W.C. Evaluation and Optimization of DNA Extraction and Purification Procedures for Soil and Sediment Samples. Appl. Environ. Microbiol. 1999, 65, 4715–4724. [Google Scholar] [CrossRef]
- Gray, J.P.; Herwig, R.P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 1996, 62, 4049–4059. [Google Scholar] [CrossRef]
- Plassart, P.; Terrat, S.; Thomson, B.; Griffiths, R.; Dequiedt, S.; Lelievre, M.; Regnier, T.; Nowak, V.; Bailey, M.; Lemanceau, P.; et al. Evaluation of the ISO Standard 11063 DNA Extraction Procedure for Assessing Soil Microbial Abundance and Community Structure. PLoS ONE 2012, 7, e44279. [Google Scholar] [CrossRef]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Moré, M.I.; Herrick, J.B.; Silva, M.C.; Ghiorse, W.C.; Madsen, E.L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 1994, 60, 1572–1580. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerra, V.; Beule, L.; Lehtsaar, E.; Liao, H.-L.; Karlovsky, P. Improved Protocol for DNA Extraction from Subsoils Using Phosphate Lysis Buffer. Microorganisms 2020, 8, 532. https://doi.org/10.3390/microorganisms8040532
Guerra V, Beule L, Lehtsaar E, Liao H-L, Karlovsky P. Improved Protocol for DNA Extraction from Subsoils Using Phosphate Lysis Buffer. Microorganisms. 2020; 8(4):532. https://doi.org/10.3390/microorganisms8040532
Chicago/Turabian StyleGuerra, Victor, Lukas Beule, Ena Lehtsaar, Hui-Ling Liao, and Petr Karlovsky. 2020. "Improved Protocol for DNA Extraction from Subsoils Using Phosphate Lysis Buffer" Microorganisms 8, no. 4: 532. https://doi.org/10.3390/microorganisms8040532
APA StyleGuerra, V., Beule, L., Lehtsaar, E., Liao, H.-L., & Karlovsky, P. (2020). Improved Protocol for DNA Extraction from Subsoils Using Phosphate Lysis Buffer. Microorganisms, 8(4), 532. https://doi.org/10.3390/microorganisms8040532







