Abstract
Lotus spp. are widely used as a forage to improve pastures, and inoculation with elite rhizobial strains is a common practice in many countries. However, only a few Lotus species have been studied in the context of plant-rhizobia interactions. In this study, forty highly diverse bacterial strains were isolated from root nodules of wild Lotus parviflorus plants growing in two field locations in Portugal. However, only 10% of these isolates could nodulate one or more legume hosts tested, whereas 90% were thought to be opportunistic nodule associated bacteria. Phylogenetic studies place the nodulating isolates within the Bradyrhizobium genus, which is closely related to B. canariense and other Bradyrhizobium sp. strains isolated from genistoid legumes and Ornithopus spp. Symbiotic nodC and nifH gene phylogenies were fully consistent with the taxonomic assignment and host range. The non-nodulating bacteria isolated were alpha- (Rhizobium/Agrobacterium), beta- (Massilia) and gamma-proteobacteria (Pseudomonas, Lysobacter, Luteibacter, Stenotrophomonas and Rahnella), as well as some bacteroidetes from genera Sphingobacterium and Mucilaginibacter. Some of these nodule-associated bacteria expressed plant growth promotion (PGP) traits, such as production of lytic enzymes, antagonistic activity against phytopathogens, phosphate solubilization, or siderophore production. This argues for a potential beneficial role of these L. parviflorus nodule-associated bacteria.
1. Introduction
Lotus is the largest genus of the Loteae tribe, which includes more than 180 species of perennial and annual herbs and small shrubs distributed in the Mediterranean region and in the Western USA [1,2]. Several Lotus spp. (Lotus uliginosus, Lotus corniculatus, Lotus tenuis, and Lotus subbiflorus) have forage value and are widely used to improve pastures [2]. Moreover, L. japonicus has been chosen as a model legume for genetic studies [3,4]. Like most legumes, Lotus spp. can establish nitrogen-fixing symbioses with certain soil bacteria collectively known as rhizobia. Typically, Lotus can either form specific interactions with the intermediate growing Mesorhizobium genus (group I: L. corniculatus and L. tenuis, e.g., [5,6,7]) or with the slow growing, Bradyrhizobium genus (group II: L. uliginosus (Lotus pedunculatus), Lotus angustissimus and L. subbiflorus, e.g., [8,9,10]). Nevertheless, Lotus-rhizobia interactions seem to be more complex, as Lotus from group I have been reported to interact with rhizobia belonging to the Rhizobium or Aminobacter genera [11]. Additionally, it has been found that certain non-cultivated Lotus species interact with rhizobia of the genus Ensifer [12,13,14]. Therefore, it seems that a lot is to be discovered about Lotus root nodule microsymbionts, especially regarding uncultivated species [15].
Genistoid legumes (e.g., Lupinus spp., Cytisus spp, Adenocarpus spp., Genista spp., Spartium ssp, etc.), Serradella (Ornithopus spp.), and L. uliginosus have been shown to be nodulated by closely related bradyrhizobia that are in the same cross-inoculation group [9,16,17]. The genus Bradyrhizobium comprises highly diversified bacteria, with 39 described species thus far [18,19,20], including some photosynthetic strains [21,22,23]. The 16S rRNA gene is very conserved within this genus and, like the symbiotic genes, has been shown to be laterally transmitted [24,25]. Thus, for a better phylogenetic prediction, multilocus sequence analysis (MLSA) of protein encoding housekeeping genes, such as recA, atpD and glnII, is more suitable [16,24,25,26].
The legume root nodules have been traditionally considered to be exclusively colonized by rhizobial bacteria, but this view has rapidly changed over the past several years. In addition to rhizobia, other bacteria can frequently inhabit the interior of nodules, e.g., Bacillus, Pseudomonas, Agrobacterium, and others [27,28,29,30,31,32,33], without causing any damage or disease, and are thus considered nodule endophytic bacteria. These nodule inhabitants are not able to nodulate by themselves and therefore could be viewed as opportunistic colonizers [34]. Recently, it has been reported that L. japonicus can control infection of nodules by both the symbiotic and other endophytic bacteria [34]. These findings have given a new perspective for these endophytic bacteria, originally treated as “contaminants”. The role of these non-rhizobia nodule bacteria is still unclear, but some studies point out a possible beneficial role, e.g., enhancing nodulation and nitrogen fixation [35,36,37]. Moreover, besides nitrogen fixation, plant growth can be enhanced through other mechanisms by rhizobia and non-rhizobial bacteria, generally known as plant growth promoting rhizobacteria (PGPR) [38] or plant probiotic bacteria (PPB). Among these other mechanisms are solubilization of mineral nutrients [39,40,41], production of phytohormones [42,43,44], or the expression of antagonistic activities against phytopathogens [45,46,47].
The aim of this work was to identify and characterize bacteria inhabiting the interior of root nodules of field-grown Lotus parviflorus plants. To our knowledge, this is the first report on bacteria isolated from root nodules from this legume.
2. Materials and Methods
2.1. Isolation of Bacterial Strains and Growth Conditions
Bacteria were isolated from effective root nodules of wild L. parviflorus plants growing in two field sites in Portugal: Beduído, which belongs to Estarreja city (site A; 40°46′0.064″ N 8° 35′12.035″ W); and fields of the “Companhia das Lezírias” farming exploration company, located in Samora Correia, which belongs to Benavente city (site B; 38° 49′24.7″ N 8°50′36.8″ W; Figure 1). Nodules were surface sterilized [9] then individually crushed before a droplet was spread on a plate with yeast-mannitol agar (YMA) supplemented with congo red dye according to [48,49]. Plates were incubated at 28 °C in the dark for 1 week. The isolate’s purity was checked by examining colony morphology and congo red absorption. Sub-culturing was conducted when more than one type of colony was present. Each pure isolate was stored at 4 °C and at −80 °C with 20% of glycerol.
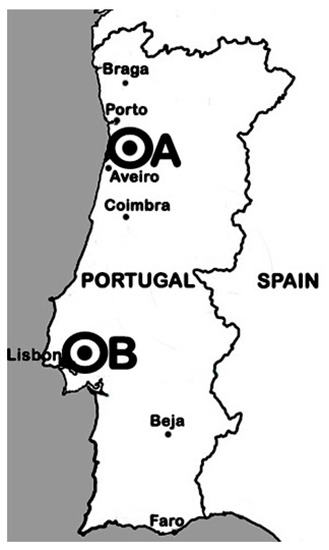
Figure 1.
Map location of the sampling sites (A and B).
Absorption of congo red was visualized by growing bacteria in YMA supplemented with 0.25 mg L−1 congo red. Congo red dye is often used to distinguish rhizobia from non-rhizobial strains. Rhizobium colonies typically do not absorb congo red [48,49], while other bacteria absorb it.
2.2. Repetitive Extragenic Palindromic (REP)-PCR Amplification and Fingerprint Analysis
Genomic DNA extractions were done following the procedure by Estrella et al. [11] or using “Aqua pure Genomic DNA extraction” kit from Bio-Rad, following the kit protocol specifications. After DNA extraction, genomic repetitive extragenic palindromic (REP) fingerprints were generated using REP1R and REP2I primers, as previously described by Versalovic, Koeuth, and Lupski [50]. Computer-assisted analysis of the REP fingerprints was performed using GelJ v2.0 software [51]. A dendrogram was constructed using dice coefficient and the unweighted pair group method with arithmetic mean method (UPGMA) clustering method [52] at a 1% band-position tolerance level.
2.3. PCR Gene Amplification and Sequencing
16S rRNA gene amplification was performed using 41F and 1488R primers as described by [53] or with primers rD1 and fD1 as described by Herrera-Cervera et al. [54]. recA and atpD gene fragments were amplified following Gaunt et al.’s methods [55]. For recA gene amplification, recA6, recA63, recA504, and recA555 primers were used; for atpD gene amplification, atpD273, atpD294 and atpD771 primers were used. glnII gene was amplified using glnR and glnF primers [56]. nodC and nifH genes were amplified following Laguerre et al.’s methods [57], using primers nodCFn, nodCF2, nodCF4, and nodCFu, nodCI, nifHF, and nifHR primers, respectively. Newly designed primers (Table 1) were used when amplification with the previous primers was not possible. For these PCR reactions primers were used at a concentration of 5 μM, together with 15.8 μL of Qiagen kit Taq mix solution (2.5 U of taq polymerase, 1.5 mM of MgCl2, 200 μM of the different dNTP), and approximately 20 ng of genomic DNA, in a final volume of 20 μL. Amplifications were performed using Eppendorf Mastercyler Gradient thermocycler. For recA amplification, samples were incubated for 5 min at 95 °C followed by 35 rounds of thermocycling conditions (95 °C for 45 s, 59 °C for 1 min, and 72 °C for 1 min 30 s) and a final extension phase (72 °C, 7 min). For atpD, samples were incubated for 5 min at 95 °C followed by 35 rounds of thermocycling conditions (95 °C for 45 s, 53 °C for 1 min and 72 °C for 2 min, using atpD183/atpD872 primers, or 95 °C for 45 s, 58 °C for 1 min, and 72 °C for 2 min, using atpD133/atpD762 primers) and a final extension phase (72 °C for 7 min). Primer 16SIR was only used for fully sequencing the 16S amplicon from some strains. The amplified DNA fragments were electrophoresed and extracted from agarose with the QIAEXII kit (Qiagen; Venlo, Netherlands) and sequenced with an ABI373 automated sequencer in the sequencing service of the “Instituto de Parasitología y Biomedicina, Lopez Neyra” CSIC, Spain. Alternatively, PCR products were sequenced using an ABI 3730 XL automated sequencer, by Stabvida, Caparica, Portugal.

Table 1.
Newly designed primers for PCR and sequencing.
2.4. NCBI Accession Numbers
Sequences obtained previously were deposited in the NCBI database. Below a list of all accession codes is presented next to the correspondent strain, for each gene.
- 16S rRNALpA5a: MK611706; LpA5b: MK611741; LpA6: MK611742; LpA7b: MK611744; LpA8: MK611745; LpA9: MK611746; LpA10: MK611747; LpA11: MK611748; LpA12: MK611749; LpA13a: MK611750; LpB5b: MT071928; LpB5d: MT071934; LpB9a: MT071930; LpB10a: MT071937; LpB10b: MT071935; LpB10c: MT071938; LpB10d: MT071936; LpB12a: MT071929; LpB12b: MT071924; LpB13: MT071932; LpB14a: MT071927; LpB14b: MT071931; LpB15b: MT071926; LpB16c: MT071939; LpB16d: MT071925; LpB17b: MT071933; LpB19a: MK611753; LpB23b: MK611754; LpB26: MK611755.
- RecALpA5a: MK671539; LpA5b: MK690337; LpA6: MK671540; LpA7b: MK671541; LpA8: MK690338; LpA9: MK671542; LpA11: MK671543; LpA13a: MK690339; LpB19a: MK671547; LpB23b: MK671548; LpB26: MK671549.
- AtpDLpA5a: MK671550; LpA5b: MK690340; LpA6: MK671551; LpA7b: MK671553; LpA8: MK690341; LpA9: MK671554; LpA10: MK690342; LpA11: MK671555; LpA13a: MK671557; LpB19a: MK671560; LpB23b: MK671561; LpB26: MK671562.
- GlnIILpA8: MK690343; LpA9: MK690344; LpA11: MK671563; LpA13b: MK690346; LpA14: MK690347; LpB19a: MK671564; LpB23b: MK671565; LpB26: MK671566.
- NodCLpA11: MK671567; LpB19a: MK671568; LpB23b: MK671569; LpB26: MK671570.
- NifHLpA11: MK690348; LpB19a: MK671571; LpB23b: MK671572; LpB26: MK671573.
2.5. Phylogenetic Analysis
Nucleotide sequences were visualized in a chromatogram and edited manually, as necessary, with Chromas lite program (version 2.1.1). Homologous sequences in NCBI Genebank database were identified using BLASTn tool. Alignments were performed using ClustalW and phylogenetic reconstruction was performed using IQ-tree [58] on XSEDE [59] and CIPRES platforms [60], with the Maximum Likelihood (ML), ModelFinder [61], and Ultra-Fast bootstrap [62] methods. An appropriate outgroup was selected to perform the ML method. The bootstrap confidence values were based on 1000 replications. Tree visualization was performed using the Interactive Tree Of Life tool [63].
2.6. Nodulation Tests
The nodulation ability of isolates was tested in the original host L. parviflorus (seeds collected from location B), as well as in related legumes: L. uliginosus cv. Sunrise; L. tenuis cv. Pampa INTA; L. tenuis cv. Esmeralda; L. corniculatus cv. San Gabriel; Lupinus luteus cv. Nacional; and Glycine max 104904B-Próvida. Seeds were surface sterilized according to Hahn and Somasegaran and Hoben [48,49]. After 1–2 h in sterilized water, seeds were transferred to 0.8% w v−1 agar-water plates for 1–2 (Lotus seeds) or 2–4 days (Lupinus and Glycine seeds). After germination, seedlings were transferred to slants (Lotus spp.) or to Leonard-type jars (Lupinus and Glycine) containing N-free Jensen plant nutrient medium [64]. Each isolate was inoculated by applying a 1 mL bacterial suspension (approximately 109 bacterial cells in ¼ Jensen medium), on the roots of each seedling (3 replicates per each bacteria–host combination). Additionally, controls (TN) fertilized with 1 mL (Lotus spp.) or 2 mL (Lupinus and Glycine) of 4.45 mM KNO3− (final concentration), and non-inoculated (T0) supplemented with 1 mL of N-free liquid Jensen medium (¼ diluted), were also prepared. Plants were grown in environmental chambers under controlled conditions (16/8 h photoperiod and 23/18 °C). Lupinus and Glycine plants were transferred to the greenhouse after 4 weeks of growth in the environmental chambers. After 8 weeks of growth, the ability to nodulate and fix nitrogen was evaluated by visual inspection of the nodules, and by comparison of plant sizes and aspect with T0 and TN controls. In addition, dry weights of aerial biomass were measured, after drying at 80 °C for 2 days, to determine the relative symbiotic effectiveness, as described by Ferreira and Marques [65].
2.7. Tests for Plant-Growth-Promotion (PGP) Traits
Qualitative tests for classical PGP activities, such as phosphate (P) solubilization, siderophore production, antagonism against phytopathogens, and production of lytic enzymes (pectinases and celullases) were carried out. Solubilization of tricalcium phosphate was tested in YED-agar plates supplemented with 0.2% of Ca3PO4 [66]. Isolates with halos around the colonies were considered as P solubilizers. The presence of siderophores was tested in TY media [67] using a CAS solution, which contains iron conjugated with a dye as a color indicator, as described by Pérez-Miranda et al. [68]. Isolates that formed orange halos were considered siderophore producers. For the antagonistic activity against phytopathogens, a portion of Phytophthora cinnamomi or Botryosphaeria cortícola mycelium was inoculated in the center of YMA plates. After 2 days of incubation at 27 °C in the dark, the bacterial isolates were inoculated in the periphery of the plate and incubated for 10 days at 27 °C in the dark. The antagonistic activity was considered when inhibition of the phytopathogens growth was visualized. Production of cellulases or pectinases were tested in TY media supplemented with 0.2% w v−1 of carboxymethylcellulose (CMC) or 0.5% w v−1 of Pectin, following procedures by Verma, Ladha, and Tripathi [69]. The appearance of halos around the colonies was accounted for the presence of the respective lytic enzymes (cellulases or pectinases).
3. Results
3.1. Bacterial Diversity
A total of 13 isolates from site A (named LpA) and 27 from site B (named LpB) were obtained from the root nodules of L. parviflorus plants. In some instances, more than one colony type (colony morphology, distinct congo red absorption, etc.) coming from a single nodule were visualized, (named types a, b, c, etc.; e.g., LpA5a and LpA5b). A broad range of congo red staining levels and growing velocity were observed among the isolated colonies (Table 2).

Table 2.
Congo red absorption and growth speed of bacteria isolated from L. parviflorus nodules.
The majority of the isolates were fast or very fast growers and did not absorb the congo red stain. The only isolates that were slow growers and did not absorb congo red were LpA11 from site A and LpB19a, LpB23b, and LpB26 from site B. The appearance of visible colonies within 24 h that also absorbed congo red (Table 2) was an indication that some bacteria might not be rhizobia [49].
REP fingerprints were obtained for all isolates except for four (LpB5c, LpB5e, LpB9b, and LpB23a). Fingerprints were composed of 2–18 bands with sizes between 200–8.000 bp (Figure S1). Considering a similarity index of 80% for strain differentiation [16], the analysis revealed 29 different clusters among the 36 isolates tested. In several instances, isolates obtained from the same nodule that had been considered visually different still displayed identical fingerprints (e.g., LpA7a and LpA7b; LpB5b, LpB5g, and LpB5f; LpB16a, LpB16b, and LpB16d). In other instances, identical fingerprints were shown by isolates originating from different nodules (LpA12, LpA13b, and LpA14; Figure S1).
3.2. Phylogenetic Analyses
3.2.1. 16S rRNA Gene
A single 1500 bp fragment, corresponding to almost full-length 16S rRNA gene, was PCR amplified of one isolate from each cluster (29 in Total; Figure S1) and further sequenced.
Aligned sequences of 920 nt positions were used to construct the phylogenetic ML tree shown in Figure 2, where four major clades can be observed. Isolates LpB16c and LpB16d were not included in this tree because only small sequence stretches from the 16S rRNA gene could be obtained. Nonetheless, highest sequence identities obtained were still included in the tree and are represented in bold and grey.
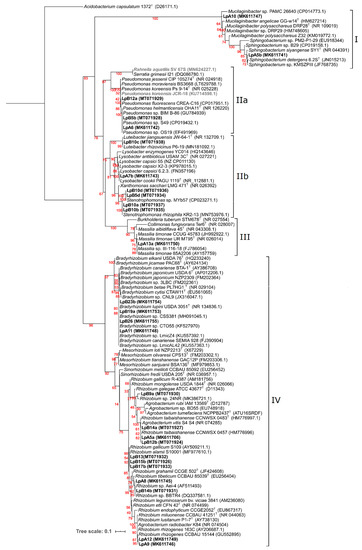
Figure 2.
Phylogeny of 16S rRNA based on 920 positions. Analyses were conducted with IQ-tree using Maximum Likelihood (ML) and ModelFinder (Best model: TIM3+F+I+G4) methods. Confidence levels of bootstrap are presented in red near each node. NCBI Genebank accession codes are presented next to each strain. Acidobacterium capsulatum 1372T (D26171.1), a bacterium from the Acidobacteria phylum, was selected as an outgroup. Isolates obtained from L. parviflorus nodules are in bold.
Clade I included bacteria belonging to the Bacteroidetes phylum (Sphingobacteriaceae) family. Isolates LpA5b and LpA10 were placed in this clade, and their closest relatives were Sphingobacterium sp. KMSZPIII and Mucilaginibacter sp. PAMC 26640, respectively. Clade II contained bacteria from the gamma-proteobacteria class. Pseudomonadaceae and Enterobacteriaceae families comprised subclade IIa, where LpA6, LpB5b, and LpB12a were included. Isolates LpA6 and LpB5b were grouped with Pseudomonas helmanticensis OHA11 and other unclassified Pseudomonas spp. The closest relative of LpB12a was Pseudomonas fluorescens CREA-C16. LpB16d and LpB16c (not included in this tree) would have the highest identity with Pseudomonas koreensis JCR-18 (100% identity) and Rahnella aquatilis SV_67S (100% identity), respectively. Subclade IIb was composed by Rhodanobacteraceae and Xanthomonadaceae families. Isolate LpB10c was grouped with Luteibacter rhizovicinus P6-19. Isolates LpB5d and LpB10d were clustered together and with Stenotrophomonas sp. MYb57. Isolates LpB10a and LpB10b were clustered together and with Stenotrophomonas rhizophila KR2-13. Isolate LpA7b was clustered with Lysobacter cookii PAGU 1119 and L. capsici 6.2.3. Clade III included bacteria from the beta-proteobacteria class (Oxalobacteraceae family). LpA13a was the only isolate placed in this clade and was grouped with Massilia timonae 85A2206 and Massilia sp. II-116-18. Clade IV comprised of bacteria belonging to the alpha-proteobacteria class (Bradyrhizobiaceae Rhizobiaceae and Phyllobacteriaceae families). Isolates LpA11, LpB19a, LpB23b, and LpB26 were clustered with Bradyrhizobium canariense SEMIA 928, Bradyrhizobium lupini USDA 3051, and other Bradyrhizobium strains isolated from genistoid legumes. Isolate LpB9a was clustered with Rhizobium sp. 24NR. Isolates LpA5a, LpB14a, and LpB12b were clustered in a group with Rhizobium taibaishanense CCNWSX 0457. LpB13, LpB15b, and LpB17b were clustered together and with Rhizobium gallicum S109 and Rhizobium alamii S10001. Isolate LpB14b was grouped with Rhizobium sp. BBTR4, while LpA8 was grouped isolated in a sister cluster. Isolates LpA9 and LpA12 were clustered in a separated group close to Rhizobium rhizogenes CCBAU 15144
3.2.2. Protein-Encoding Housekeeping Genes
Since 16S rRNA gene cannot provide the best phylogenetic resolution for some bacteria, like in the case of Bradyrhizobia, MLSA of several protein-encoding housekeeping genes was performed. Only site B strains that nodulated (LpB19a, LpB23b, and LpB26; Table 3) were considered for further phylogenetic analysis, while all distinctive isolates by REP-PCR (80% similarity) and 16S rRNA gene sequence from site A (LpA5a, LpA5b, LpA6, LpA7b, LpA8, LpA9, LpA10, LpA11, and LpA13a) were considered. From these, only LpA8, LpA9, LpA11, LpB19a, LpB23b, and LpB26 genomic DNAs produced amplicons for all housekeeping genes tested, with sizes of approximately 500 bp for recA and atpD, and 700bp for glnII. The rest of the isolates failed mainly to produce amplicons for glnII gene, which unlike glnA is not ubiquitous in bacteria [70]. For isolates LpA5b, LpA6, LpA10, and LpA13a we could obtain an atpD amplicon of approximately 700 bp only after using the newly designed primers (atpD183 and atpD872 for LpA5b and LpA10, and atpD133 and atpD762 for LpA6 and LpA13a; see Table 1). Also, the newly designed recA primers allowed us to obtain a recA amplicon of almost 600 bp from isolate LpA5b.

Table 3.
Host range assays performed in different legume host plants inoculated with the different bacterial isolates.
The protein encoding housekeeping genes phylogeny was congruent with the 16S rRNA phylogeny at the genus and also very often at the species level. As there are few sequences available in the NCBI Genebank database for genera Sphingobacterium, Lysobacter, Mucilaginibacter, and Massilia, most of the sequences for these genera were gathered from genome sequencing projects. For the isolates from which all three housekeeping genes could be amplified, a ML phylogenetic tree was generated using concatenated sequences of glnII, atpD, and recA genes (Figure 3). The clustering of isolates LpA11, LpB19a, LpB23b, and LpB26 within Bradyrhizobium was confirmed to be closely related to B. canariense. The closest relative of LpA11 was B. canariense Oc1 (99.6% sequence identity), a strain isolated from Ornithopus compressus in Sardinia, Italy [71]. LpB19a phylogenetic position was less resolved. It was grouped in a low confidence cluster with Bradyrhizobium canariense BC-P22 (98.3% identity), a strain isolated from Chamaecytisus proliferus in the Canary Islands [72], while sharing a higher identity with Bradyrhizobium sp. LmicAL42 (99.6% identity), a strain isolated from Lupinus micranthus nodules in Spain [73]. LpB23b and LpB26 were clustered together in a highly supported cluster with Bradyrhizobium sp. LmicZ4 (100% similarity for LpB23b and 99.9% of similarity for LpB26), a strain isolated from Lupinus micranthus nodules in Algeria [73]. On the other hand, isolates LpA8 and LpA9 were again clustered within the Rhizobium/Agrobacterium group, albeit not matching any previously described species.
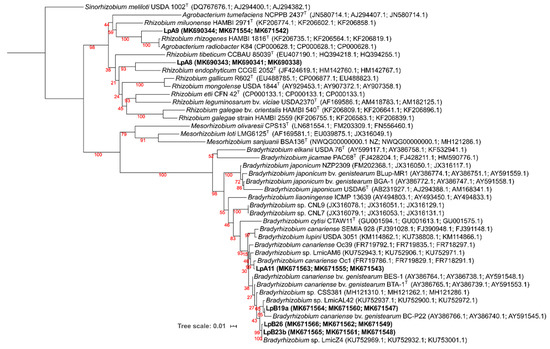
Figure 3.
Phylogeny of concatenated sequences of glnII (300 nt), atpD (328 nt), and recA (333 nt) genes, based on a total of 968 nt. Bootstrap values are shown in red at each node. Analyses were conducted with IQ-tree using Maximum Likelihood (ML) and ModelFinder (Best model: TIM+F+I+G4) methods. NCBI Genebank accession codes are shown. Sinorhizobium meliloti USDA 1002T (DQ767676.1; AJ294400.1; AJ294382.1) was selected as an outgroup. Isolates obtained from L. parviflorus nodules are in bold.
For those isolates that were negative for glnII amplification, ML phylogenetic trees of recA and atpD genes were constructed (Figure S2). Isolate LpA5a was clustered with Rhizobium taibaishanense HAMBI 3414T (now Allorhizobium taibaishanense, in accordance with Bourebaba et al. [74]), having a similarity of 98.2% in recA and 98.6% atpD gene, respectively. LpA5b was placed in a clade of Sphingobacterium spp., having Sphingobacterium sp. B29 as the closest relative (98.5% identity in the atpD gene and 92.1% identity in the recA gene). LpA6 was clustered with Pseudomonas spp., having Pseudomonas koriensis DSM 11610T (99.7% identity) and Pseudomonas jessenii DSM 17150T (96.1% identity) as the closest relatives in the atpD and recA gene trees, respectively. Isolate LpA7b was placed in the Lysobacter genus, sharing the highest identities with L. capsici 55 (99.2% in the atpD gene and 99.3% in recA gene). Isolate LpA10 was clustered in a single branch within the Mucilaginibacter clade, sharing only 85.4% and 84.8% with Mucilaginibacter mallensis MP1X4 and Mucilaginibacter sp. PAMC 26640, respectively, in the atpD gene. Lp13a was grouped with Massilia spp., sharing 100% of similarity, in both genes, with M. timonae CCUG 45783.
3.2.3. Symbiotic Genes: nodC and nifH
Symbiotic nodC and nifH gene amplicons were obtained only from strains LpA11, LpB19a, LpB23b, and LpB26, which matched their nodulation ability (Table 3). For both gene phylogenies (Figure 4 and Figure 5), these isolates were clustered in a well-supported clade, comprising of B. canariense and other Bradyrhizobium sp. strains isolated from genistoid legumes. The closest Lotus rhizobia were isolates from L. uliginosus, which is in line with the results of the nodulation tests (Table 3).

Figure 4.
Phylogenetic tree of nodC gene sequences. Analyses were conducted with IQ-tree using Maximum Likelihood (ML) and ModelFinder (Best model: HKY+F+G4) methods), for a total of 203 positions. Bootstrap values are shown in red at each node. NCBI Genebank accession codes and original legume host of isolation are indicated. Burkholderia tuberum STM678T (AJ306730.1), a bacterium from the beta-proteobacteria class that nodulates Aspalathus carnosa, was selected as an outgroup. Lotus spp. and isolates obtained from L. parviflorus nodules are in bold.
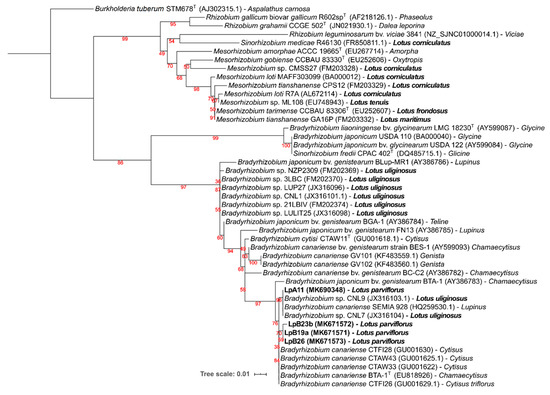
Figure 5.
Phylogenetic tree of nifH gene sequences. Analyses were conducted with IQ-tree using Maximum Likelihood (ML) and ModelFinder (Best model: K2P+G4) methods), for a total of 203 positions. Bootstrap values are shown at each node. NCBI Genebank accession codes and original legume host of isolation are indicated. Burkholderia tuberum STM678T (AJ302315.1), a bacterium from the beta-proteobacteria class that nodulates Aspalathus carnosa, was selected as an outgroup. Lotus spp. and isolates obtained from L. parviflorus nodules are in bold.
3.3. Nodulation Tests
Symbiotic ability (nodulation and effectiveness) was tested for all isolates in various Lotus species. Isolates that nodulated Lotus plants were additionally tested in Lupinus luteus and Glycine max plants (Table 3). Only isolates LpA11 from sampling site A, and LpB19a, LpB23b, and LpB26 from site B were able to nodulate any of the Lotus spp. tested. LpA11 could nodulate and fix nitrogen in L. uliginosus, L. parviflorus, and L. luteus. It also formed non-fixing nodules in L. corniculatus and L. tenuis cv. Esmeralda, as judged from nodule appearance (white color) and plants dry weight. No nodulation was observed in L. tenuis cv. INTA PAMPA, and G. max. LpB19a, LpB23b, and LpB26 isolates displayed a Nod+ Fix+ phenotype in L. uliginosus, L. parviflorus, and L. luteus, as well as a Nod+Fix− phenotype in L. corniculatus and L. tenuis cv. INTA PAMPA. Similar to LpA11, none of them formed nodules in G. max. The results of the symbiotic tests are in accordance with the symbiotic genes phylogenies, that placed the L. parviflorus symbionts close to rhizobia from genistoid legumes and L. uliginosus, but distant from other Lotus spp. and Glycine rhizobia (see Figure 4 and Figure 5).
3.4. PGP Traits
Nearly 40% of the non-nodulating isolates displayed one or more activities related with plant growth promotion or protection (Table 4; Figures S3 and S4). Particularly abundant was the production of cellulases, often by strains that also showed antagonistic activity against fungal phytopathogens (isolates LpA7a, LpA7b, LpB16a, LpB16b, LpB16d, and LpB23a). In contrast, only one isolate (LpB16d) showed significant P solubilization capacity, whereas 4 isolates (LpA6, LpB12a, LpB5b, and LpB5c) showed siderophore production under our experimental conditions (Table 4). These results, together with the phylogenetic analyses described above, indicate that the non-nodulating bacteria isolated from L. parviflorus nodules are common soil and rhizosphere inhabitants, which can eventually exhibit plant beneficial traits. For instance, isolates LpA7a and LpB16d, which showed antagonistic activity against phytopathogens and lytic enzyme production (Table 4), are closely related to Lysobacter capsici and Pseudomonas koreensis, respectively. These species are well known for a suppressive activity on several plant pathogens [46,75,76,77,78]. Moreover, isolates LpA6, LpB5b, LpB12a, and LpB16d, which showed siderophore production, are closely related to Pseudomonas spp. (Pseudomonas koreensis and Pseudomonas fluorescens) that are commonly reported to show this trait [77,79,80,81]. Similarly, phosphate solubilization was shown by the pseudomonad isolate LpB16d that is related to P. koreensis, which has also been shown previously to solubilize phosphate [82,83,84].

Table 4.
Plant growth promotion (PGP) traits: Phosphate solubilization, siderophore production, antagonistic activity against phytophatogens, and lytic enzymes production. Only isolates that showed at least one activity assayed are presented, even though all isolates were tested.
4. Discussion
We found a great diversity of bacteria inside nodules of wild L. parviflorus plants growing in two different locations in Portugal. Intriguingly, only four of these isolates (10%) were able to nodulate the original host L. parviflorus. These four isolates are slow growers, and phylogenetically related to B. canariense and other Bradyrhizobium strains isolated from legumes, such as L. uliginosus, Ornithopus and other genistoid legumes (e.g., Lupinus) root nodules. Indeed, L. parviflorus bradyrhizobia were also able to effectively nodulate L. uliginosus and L. luteus. The other 30 isolates were not able to nodulate any of the legume hosts tested. Often, the symbiotic bradyrhizobia and the non-nodulating bacteria were isolated from the same nodule, suggesting that the later could be opportunistic nodule inhabitants. Almost all these non-nodulating bacteria displayed faster growth rates than the nodulating bradyhrizobia, which would explain why they were much more frequently isolated than the actual symbiotic bradyrhizobia. However, we have isolated slow-growing bradyrhizobia from other related legumes, such as L. uliginosus [9,16], following similar procedures, without observing such an abundance of fast-growing bacteria inside the nodules. Thus, for some unknown reason the nodules of L. parviflorus seem more frequently invaded by non-nodulating opportunistic bacteria than other Lotus spp. Alternatively, other legume nodule endophytes may be unculturable or unable to grow in the YMA medium generally used for rhizobial isolation. Nevertheless, the high abundance of fast-growing nodule endophytes has seriously hindered our goal of describing the genetic diversity among L. parviflorus rhizobia. The presence of these fast-growing strains when cultivated could inhibit the growth of the bradyrhizobia that are able to nodulate L. parviflorus. Fast growers are able to spread on the agar growth media, likely disturbing or even inhibiting the growth of bradyrhizobia. In addition, some of the isolated strains could display active motility in solid surfaces (swarming, gliding, sliding), e.g., Lysobacter spp. possess gliding motility [46], that can further facilitate their spreading on the agar surface. For future studies, approaches to favor either the selective isolation of the bradyrhizobia or the counterselection of other nodule bacteria, should be implemented.
Besides nitrogen fixation, some authors also show other PGP activities from rhizobia, such as phosphate solubilization, siderophore production, and biocontrol activity [40,85,86,87]. However, we did not find any PGP activity (besides nitrogen fixation) from the nodulating bradyrhizobia, and only one non-nodulating Rhizobium spp. strain (LpB12b) showed lytic enzyme production. Indeed, the use of co-inoculations of rhizobia and other bacteria more specialized for other PGP traits is a common practice in biotechnological applications in agriculture [88,89,90,91]. All L. parviflorus nodule bacteria were related to previously described soil or plant-associated taxa. We observed that many of these nodule inhabitants could exhibit activities potentially beneficial for plant growth and health, including phosphate solubilization, production of siderophores and lytic enzymes, as well as antagonism against two well-known phytopathogens, Phytophthora cinnamomi and Botryosphaeria cortícola. Endophytic bacteria with these beneficial activities are frequently found in legume tissue, including nodules [92,93,94]. The isolation of bacteria other than rhizobia from the interior of the legume nodules has been increasingly reported in precious years [28,30,31,32,33,95]. Some plant species seem more prone to harbor a high amounts and diversity of non-rhizobia inside their nodules, as reported by De Meyer et al. [28], who studied 654 isolates from the nodules of 30 different plant species. Moreover, there are certain endophytic bacteria that have been frequently isolated from legume root nodules, e.g., Micromonospora sp. [33], Bacillus sp., and Pseudomonas sp. [28]. Remarkably, [34] have shed some light in the understanding of this phenomenon. By following the progression of L. japonicus nodule infection and colonization by M. japonicum R7A and endophytic strains, these authors found that the endophyte’s ability to infect and colonize nodules was discriminated and controlled by the plant. Exopolysaccharides produced by endophytic bacteria and early symbiotic plant genes play a vital role to enable and favor their nodule invasion and colonization through infection threats initiated by M. japonicum R7A. This determinant molecular crosstalk between endophytes and plants could explain why some legume plants are more prone to be colonized by certain endophytes. The fact that we isolated closely related bacteria from two different locations separated by nearly 200 Km (see Figure 2, isolates LpA5a, LpB12b, and LpB14a; LpA6 and LpB5b; LpA8 and LpB14b) argues for a potential selectivity by the host plant. In our hands, L. parviflorus nodules seem more predisposed to be colonized by non-rhizobial bacteria than other related species such as L. uliginosus [9,15,28]. Likewise, L. corniculatus has also been reported to have nodules frequently colonized by endophytic bacteria [17,96,97], albeit not to the extent we have found for L. parviflorus in this study.
It would be interesting to know if any of L. parviflorus nodule bacteria can also be found inside other plant organs, and whether there is some type of selectivity or favoritism towards certain taxa. In contrast with legume–rhizobia symbiosis, interactions of legumes with other endophytes are just starting to be unraveled. Our results reinforce the notion that legume root nodules can host complex bacterial communities that could express nitrogen fixation and additional plant growth promoting activities.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/4/499/s1, Figure S1: UPGMA dendrogram, Figure S2: Phylogeny of atpD and recA genes, Figure S3: Inhibition of P. cinnamomi and B. corticola, Figure S4: In vitro plant growth promoting rhizobacteria (PGPR) activities.
Author Contributions
Conceptualization, I.V.eC., J.S., M.J.L., and R.S; Methodology, I.V.eC., J.S., M.J.L., and R.S; Investigation, R.S., J.T., and M.J.L.; Resources, I.V.eC., J.S., and E.F.; Data Curation, R.S. and M.J.L.; Writing—Original Draft Preparation, R.S.; Writing—Review and Editing, I.V.eC., J.S., and M.J.L.; Supervision, I.V.eC., J.S, M.J.L., and E.F.; Project administration, I.V.eC. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was given by the projects PRODER, PA 54970 and ALT20-45-2015-08.
Acknowledgments
This work was supported by an Erasmus grant (2012-1-PT1-ERA02-12581), and a BIOFAG (CYTED) grant to Ricardo Soares. We thank Jorge Capelo for the identification of Lotus parviflorus plants and Helena Machado and Helena Bragança for the Phytophthora cinnamomi and Botryosphaeria corticola, strains, respectively.
Conflicts of Interest
The authors declare no conflict of interest that could potentially influence the outcome of the study.
References
- Allan, G.J.; Francisco-Ortega, J.; Santos-Guerra, A.; Boerner, E.; Zimmer, E.A. Molecular phylogenetic evidence for the geographic origin and classification of Canary Island Lotus (Fabaceae: Loteae). Mol. Phylogenetics Evol. 2004, 32, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Díaz, P.; Borsani, O.; Monza, J. Lotus-related species and their agronomic importance. In Lotus japonicus Handbook; Springer-Verlag: Berlin/Heidelberg, 2005; pp. 25–37. [Google Scholar]
- Márquez, A. Lotus Japonicus, A General Introduction; Springer: Sevilha, Spain, 2005; pp. 3–15. ISBN 978-1-4020-3735-1. [Google Scholar]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008, 15, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Monza, J.; Fabiano, E.; Arias, A. Characterization of an indigenous population of rhizobia nodulating Lotus corniculatus. Soil Biol. Biochem. 1992, 24, 241–247. [Google Scholar] [CrossRef]
- Pankhurst, C.E.; Craig, A.S.; Jones, W.T. Effectiveness of Lotus Root Nodules: I. morphology and flavolan content of nodules formed on Lotus pedunculatus by fast-growing Lotus rhizobia. J. Exp. Bot. 1979, 30, 1085–1093. [Google Scholar] [CrossRef]
- Lorite, M.J.; Muñoz, S.; Olivares, J.; Soto, M.J.; Sanjuán, J. Characterization of strains unlike Mesorhizobium loti that nodulate lotus spp. in saline soils of Granada, Spain. Appl. Environ. Microbiol. 2010, 76, 4019–4026. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, P.; Milnitsky, F.; Monza, J.; Bedmar, E.J. Characterization of rhizobia nodulating Lotus subbiflorus from Uruguayan soils. Plant Soil 1996, 180, 39–47. [Google Scholar] [CrossRef]
- Lorite, M.J.; Videira e Castro, I.; Muñoz, S.; Sanjuán, J. Phylogenetic relationship of Lotus uliginosus symbionts with bradyrhizobia nodulating genistoid legumes. FEMS Microbiol. Ecol. 2012, 79, 454–464. [Google Scholar] [CrossRef]
- Cooper, J.E.; Wood, M.; Bjourson, A.J. Nodulation of Lotus pedunculatus in acid rooting solution by fast- and slow-growing rhizobia. Soil Biol. Biochem. 1985, 17, 487–492. [Google Scholar] [CrossRef]
- Estrella, M.J.; Muñoz, S.; Soto, M.J.; Ruiz, O.; Sanjuán, J. Genetic diversity and host range of rhizobia nodulating Lotus tenuis in typical soils of the Salado River Basin (Argentina). Appl. Environ. Microbiol. 2009, 75, 1088–1098. [Google Scholar] [CrossRef]
- Lorite, M.J.; Donate-Correa, J.; del Arco-Aguilar, M.; Pérez Galdona, R.; Sanjuán, J.; León-Barrios, M. Lotus endemic to the Canary Islands are nodulated by diverse and novel rhizobial species and symbiotypes. Syst. Appl. Microbiol. 2010, 33, 282–290. [Google Scholar] [CrossRef]
- León-Barrios, M.; Lorite, M.J.; Donate-Correa, J.; Sanjuán, J. Ensifer meliloti bv. lancerottense establishes nitrogen-fixing symbiosis with Lotus endemic to the Canary Islands and shows distinctive symbiotic genotypes and host range. Syst. Appl. Microbiol. 2009, 32, 413–420. [Google Scholar]
- Merabet, C.; Martens, M.; Mahdhi, M.; Zakhia, F.; Sy, A.; Le Roux, C.; Domergue, O.; Coopman, R.; Bekki, A.; Mars, M.; et al. Multilocus sequence analysis of root nodule isolates from Lotus arabicus (Senegal), Lotus creticus, Argyrolobium uniflorum and Medicago sativa (Tunisia) and description of Ensifer numidicus sp. nov. and Ensifer garamanticus sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Lorite, M.J.; Estrella, M.J.; Escaray, F.J.; Sannazzaro, A.; Videira e Castro, I.M.; Monza, J.; Sanjuán, J.; León-Barrios, M. The Rhizobia-Lotus Symbioses: Deeply Specific and Widely Diverse. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Batista, L.; Tomasco, I.; Lorite, M.J.; Sanjuán, J.; Monza, J. Diversity and phylogeny of rhizobial strains isolated from Lotus uliginosus grown in Uruguayan soils. Appl. Soil Ecol. 2013, 66, 19–28. [Google Scholar] [CrossRef]
- De Meyer, S.E.; Van Hoorde, K.; Vekeman, B.; Braeckman, T.; Willems, A. Genetic diversity of rhizobia associated with indigenous legumes in different regions of Flanders (Belgium). Soil Biol. Biochem. 2011, 43, 2384–2396. [Google Scholar] [CrossRef]
- Avontuur, J.; Palmer, M.; Beukes, C.Y.; Chan, W.; Coetzee, M.; Blom, J.; Stępkowski, T.; Kyrpides, N.C.; Woyke, T.; Shapiro, N.; et al. Genome-informed Bradyrhizobium taxonomy: Where to from here? Syst. Appl. Microbiol. 2019, 42, 427–439. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Martínez-Romero, E. A Genomotaxonomy View of the Bradyrhizobium Genus. Front. Microbiol. 2019, 10, 1334. [Google Scholar] [CrossRef]
- Bradyrhizobium. Available online: http://www.bacterio.net/bradyrhizobium.html (accessed on 3 September 2019).
- Evans, W.R.; Fleischman, D.E.; Calvert, H.E.; Pyati, P.V.; Alter, G.M.; Rao, N.S. Bacteriochlorophyll and Photosynthetic Reaction Centers in Rhizobium Strain BTAi 1. Appl. Environ. Microbiol. 1990, 56, 3445–3449. [Google Scholar] [CrossRef]
- Giraud, E.; Fleischman, D. Nitrogen-fixing symbiosis between photosynthetic bacteria and legumes. Photosynth. Res. 2004, 82, 115–130. [Google Scholar] [CrossRef]
- Giraud, E.; Xu, L.; Chaintreuil, C.; Gargani, D.; Gully, D.; Sadowsky, M.J. Photosynthetic Bradyrhizobium sp. strain ORS285 is capable of forming nitrogen-fixing root nodules on soybeans (Glycine max). Appl. Environ. Microbiol. 2013, 79, 2459–2462. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; Van de Peer, Y.; Vandamme, P.; Thompson, F.L.; et al. Opinion: Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005, 3, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, K.; Miyazaki, K. Revisiting bacterial phylogeny: Natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob. Genet. Elem. 2013, 3, e24210. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, O.M.; López, M.V.; Riccillo, P.M. The diversity of rhizobia nodulating beans in Northwest Argentina as a source of more efficient inoculant strains. J. Biotechnol. 2001, 91, 181–188. [Google Scholar] [CrossRef]
- De Meyer, S.E.; De Beuf, K.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef]
- Gossmann, J.A.; Markmann, K.; Brachmann, A.; Rose, L.E.; Parniske, M. Polymorphic infection and organogenesis patterns induced by a Rhizobium leguminosarum isolate from Lotus root nodules are determined by the host genotype. New Phytol. 2012, 196, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, F.; Angelini, J.; Taurian, T.; Tonelli, M.L.; Fabra, A. Endophytic occupation of peanut root nodules by opportunistic Gammaproteobacteria. Syst. Appl. Microbiol. 2009, 32, 49–55. [Google Scholar] [CrossRef]
- Kan, F.L.; Chen, Z.Y.; Wang, E.T.; Tian, C.F.; Sui, X.H.; Chen, W.X. Characterization of symbiotic and endophytic bacteria isolated from root nodules of herbaceous legumes grown in Qinghai-Tibet plateau and in other zones of China. Arch. Microbiol. 2007, 188, 103–115. [Google Scholar] [CrossRef]
- Muresu, R.; Polone, E.; Sulas, L.; Baldan, B.; Tondello, A.; Delogu, G.; Cappuccinelli, P.; Alberghini, S.; Benhizia, Y.; Benhizia, H.; et al. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 2008, 63, 383–400. [Google Scholar] [CrossRef]
- Trujillo, M.E.; Alonso-Vega, P.; Rodríguez, R.; Carro, L.; Cerda, E.; Alonso, P.; Martínez-Molina, E. The genus Micromonospora is widespread in legume root nodules: The example of Lupinus angustifolius. Isme J 2010, 4, 1265–1281. [Google Scholar] [CrossRef]
- Zgadzaj, R.; James, E.K.; Kelly, S.; Kawaharada, Y.; de Jonge, N.; Jensen, D.B.; Madsen, L.H.; Radutoiu, S. A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria. PLoS Genet. 2015, 11, e1005280. [Google Scholar] [CrossRef] [PubMed]
- Mrabet, M.; Mnasri, B.; Romdhane, S.B.; Laguerre, G.; Aouani, M.E.; Mhamdi, R. Agrobacterium strains isolated from root nodules of common bean specifically reduce nodulation by Rhizobium gallicum. Fems Microbiol. Ecol. 2006, 56, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhou, X.; Smith, D.L. Enhanced Soybean Plant Growth Resulting from Coinoculation of Strains with. Crop Sci. 2003, 43, 1774. [Google Scholar] [CrossRef]
- Tokala, R.K.; Strap, J.L.; Jung, C.M.; Crawford, D.L.; Salove, M.H.; Deobald, L.A.; Bailey, J.F.; Morra, M.J. Novel Plant-Microbe Rhizosphere Interaction Involving Streptomyces lydicus WYEC108 and the Pea Plant (Pisum sativum). Appl. Environ. Microbiol. 2002, 68, 2161–2171. [Google Scholar] [CrossRef]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Datta, B.; Chakrabartty, P.K. Siderophore biosynthesis genes of Rhizobium sp. isolated from Cicer arietinum L. 3 Biotech 2014, 4, 391–401. [Google Scholar] [CrossRef]
- Halder, A.K.; Mishra, A.K.; Bhattacharyya, P.; Chakrabartty, P.K. Solubilization of rock phosphate by Rhizobium and Bradyrhizobium. J. Gen. Appl. Microbiol. 1990, 36, 81–92. [Google Scholar] [CrossRef]
- Peix, A.; Rivas-Boyero, A.A.; Mateos, P.F.; Rodriguez-Barrueco, C.; Martínez-Molina, E.; Velazquez, E. Growth promotion of chickpea and barley by a phosphate solubilizing strain of Mesorhizobium mediterraneum under growth chamber conditions. Soil Biol. Biochem. 2001, 33, 103–110. [Google Scholar] [CrossRef]
- Datta, C.; Basu, P.S. Indole acetic acid production by a Rhizobium species from root nodules of a leguminous shrub, Cajanus cajan. Microbiol. Res. 2000, 155, 123–127. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Melentiev, A.I.; Martynenko, E.V.; Timergalina, L.N.; Arkhipova, T.N.; Shendel, G.V.; Kuz’mina, L.Y.; Dodd, I.C.; Veselov, S.Y. Cytokinin producing bacteria stimulate amino acid deposition by wheat roots. Plant Physiol. Biochem. 2014, 83, 285–291. [Google Scholar] [CrossRef]
- Malik, D.K.; Sindhu, S.S. Production of indole acetic acid by Pseudomonas sp.: Effect of coinoculation with Mesorhizobium sp. Cicer on nodulation and plant growth of chickpea (Cicer arietinum). Physiol. Mol. Biol. Plants 2011, 17, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.-L. Antagonistic activity of Rhizobium spp. against beneficial and plant pathogenic fungi. Lett. Appl. Microbiol. 1990, 10, 213–215. [Google Scholar] [CrossRef]
- Gómez Expósito, R.; Postma, J.; Raaijmakers, J.M.; De Bruijn, I. Diversity and Activity of Lysobacter Species from Disease Suppressive Soils. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.-S.; Jin, R.-D.; Krishnan, H.B.; Lee, S.-B.; Kim, K.-Y. Biocontrol ability of Lysobacter antibioticus HS124 against Phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 2009, 59, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Hahn, N.J. The Congo red reaction in bacteria and its usefulness in the identification of rhizobia. Can. J. Microbiol. 1966, 12, 725–733. [Google Scholar] [CrossRef]
- Somasegaran, P.; Hoben, H.J. Handbook for Rhizobia: Methods in legume-rhizobium technology; Springer-Verlag: Berlin, Germany, 1994; ISBN 0-387-94134-7. [Google Scholar]
- Versalovic, J.; Koeuth, T.; Lupski, J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ--a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef]
- Sneath, P.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; Freeman & Co: San Francisco, CA, USA, 1973; ISBN 0716706970 9780716706977. [Google Scholar]
- Herrera-Cervera, J.A.; Caballero-Mellado, J.; Laguerre, G.; Tichy, H.-V.; Requena, N.; Amarger, N.; MartÃ-nez-Romero, E.; Olivares, J.; Sanjuan, J. At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soil. FEMS Microbiol. Ecol. 1999, 30, 87–97. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Gaunt, M.W.; Turner, S.L.; Rigottier-Gois, L.; Lloyd-Macgilp, S.A.; Young, J.P. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 2001, 51, 2037–2048. [Google Scholar] [CrossRef]
- Stepkowski, T.; Czaplińska, M.; Miedzinska, K.; Moulin, L. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha Proteobacteria. Syst. Appl. Microbiol. 2003, 26, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, G.; Nour, S.M.; Macheret, V.; Sanjuan, J.; Drouin, P.; Amarger, N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 2001, 147, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Towns, J.; Cockerill, T.; Dahan, M.; Foster, I.; Gaither, K.; Grimshaw, A.; Hazlewood, V.; Lathrop, S.; Lifka, D.; Peterson, G.D.; et al. XSEDE: Accelerating Scientific Discovery. Comput. Sci. Eng. 2014, 16, 62–74. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); IEEE: New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Jensen, H.L. Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linn. Soc. New South Wales 1941, 67, 98–108. [Google Scholar]
- Ferreira, E.M.; Marques, J.F. Selection of Portuguese Rhizobium leguminosarum bv. trifolii strains for production of legume inoculants. Plant Soil 1992, 147, 151–158. [Google Scholar]
- Illmer, P.; Barbato, A.; Schinner, F. Solubilization of hardly-soluble AlPO4 with P-solubilizing microorganisms. Soil Biol. Biochem. 1995, 27, 265–270. [Google Scholar] [CrossRef]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Ladha, J.K.; Tripathi, A.K. Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J. Biotechnol. 2001, 91, 127–141. [Google Scholar] [CrossRef]
- Brown, J.R.; Masuchi, Y.; Robb, F.T.; Doolittle, W.F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J. Mol. Evol. 1994, 38, 566–576. [Google Scholar] [CrossRef]
- Stępkowski, T.; Zak, M.; Moulin, L.; Króliczak, J.; Golińska, B.; Narożna, D.; Safronova, V.I.; Mądrzak, C.J. Bradyrhizobium canariense and Bradyrhizobium japonicum are the two dominant rhizobium species in root nodules of lupin and serradella plants growing in Europe. Syst. Appl. Microbiol. 2011, 34, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Vinuesa, P.; Silva, C.; Werner, D.; Martínez-Romero, E. Population genetics and phylogenetic inference in bacterial molecular systematics: The roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 2005, 34, 29–54. [Google Scholar] [CrossRef]
- Bourebaba, Y.; Durán, D.; Boulila, F.; Ahnia, H.; Boulila, A.; Temprano, F.; Palacios, J.M.; Imperial, J.; Ruiz-Argüeso, T.; Rey, L. Diversity of Bradyrhizobium strains nodulating Lupinus micranthus on both sides of the Western Mediterranean: Algeria and Spain. Syst. Appl. Microbiol. 2016, 39, 266–274. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Willems, A.; Nesme, X.; de Lajudie, P.; Lindström, K. Revised phylogeny of Rhizobiaceae: Proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 2015, 38, 84–90. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, R.; Aslam, Z.; Jeon, C.O.; Chung, Y.R. Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int. J. Syst. Evol. Microbiol. 2008, 58, 387–392. [Google Scholar] [CrossRef]
- Puopolo, G.; Sonego, P.; Engelen, K.; Pertot, I. Draft Genome Sequence of Lysobacter capsici AZ78, a Bacterium Antagonistic to Plant-Pathogenic Oomycetes. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Lin, H.; Hu, S.; Liu, R.; Chen, P.; Ge, C.; Zhu, B.; Guo, L. Genome Sequence of Pseudomonas koreensis CRS05-R5, an Antagonistic Bacterium Isolated from Rice Paddy Field. Front. Microbiol 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Rafikova, G.F.; Korshunova, T.Y.; Minnebaev, L.F.; Chetverikov, S.P.; Loginov, O.N. A new bacterial strain, Pseudomonas koreensis IB-4, as a promising agent for plant pathogen biological control. Microbiology 2016, 85, 333–341. [Google Scholar] [CrossRef]
- Ganeshan, G.; Kumar, A.M. Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. J. Plant Interact. 2005, 1, 123–134. [Google Scholar] [CrossRef]
- Trapet, P.; Avoscan, L.; Klinguer, A.; Pateyron, S.; Citerne, S.; Chervin, C.; Mazurier, S.; Lemanceau, P.; Wendehenne, D.; Besson-Bard, A. The Pseudomonas fluorescens Siderophore Pyoverdine Weakens Arabidopsis thaliana Defense in Favor of Growth in Iron-Deficient Conditions. Plant Physiol. 2016, 171, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.W.; Islam, A.; Opdahl, L.; Davenport, J.R.; Sullivan, T.S. Comparative Genomics, Siderophore Production, and Iron Scavenging Potential of Root Zone Soil Bacteria Isolated from ‘Concord’ Grape Vineyards. Microb. Ecol. 2019, 78, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Gusain, Y.S.; Kamal, R.; Mehta, C.M.; Singh, U.S.; Sharma, A.K. Phosphate solubilizing and indole-3-acetic acid producing bacteria from the soil of Garhwal Himalaya aimed to improve the growth of rice. J. Env. Biol. 2015, 36, 301–307. [Google Scholar]
- Kasotia, A.; Choudhary, D.K. Induced Inorganic Phosphate Solubilization Through N-Methyl-N´-Nitro-N-Nitrosoguanidine Treated Mutants of Pseudomonas koreensis Strain AK-1 (MTCC Number 12058) under Polyethylene Glycol. Proc. Natl. Acad. Sci. USA 2016, 86, 115–123. [Google Scholar] [CrossRef]
- Kwon, S.W.; Kim, J.S.; Park, I.C.; Yoon, S.H.; Park, D.H.; Lim, C.K.; Go, S.J. Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 21–27. [Google Scholar] [CrossRef]
- Daimon, H.; Nobuta, K.; Ohe, M.; Harada, J.; Nakayama, Y. Tricalcium Phosphate Solubilization by Root Nodule Bacteria of Sesbania cannabina and Crotalaria juncea. Plant Prod. Sci. 2006, 9, 388–389. [Google Scholar] [CrossRef]
- Das, K.; Prasanna, R.; Saxena, A.K. Rhizobia: A potential biocontrol agent for soil borne fungal pathogens. Folia Microbiol. (Praha) 2017, 62, 425–435. [Google Scholar] [CrossRef]
- Deryło, M.; Choma, A.; Puchalski, B.; Suchanek, W. Siderophore activity in Rhizobium species isolated from different legumes. Acta Biochim. Pol. 1994, 41, 7–11. [Google Scholar] [CrossRef]
- Afzal, A.; Bano, A. Rhizobium and Phosphate Solubilizing Bacteria Improve the Yield and Phosphorus Uptake in Wheat (Triticum aestivum). Int. J. Agric. Biol. 2008, 10, 1560–8530. [Google Scholar]
- Ahmad, M.; Zahir, Z.A.; Khalid, M.; Nazli, F.; Arshad, M. Efficacy of Rhizobium and Pseudomonas strains to improve physiology, ionic balance and quality of mung bean under salt-affected conditions on farmer’s fields. Plant Physiol. Biochem. 2013, 63, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Berg, G.; Lindström, K.; Räsänen, L.A. Co-inoculation of Pseudomonas spp. with Rhizobium improves growth and symbiotic performance of fodder galega (Galega orientalis Lam.). Eur. J. Soil Biol. 2010, 46, 269–272. [Google Scholar] [CrossRef]
- Videira e Castro, I.; de Castro Silva, M.; Fernandez, C.; Colavolpe, B.; Machado, H. The Potential of Nitrogen-Fixing Bacteria in the Sustainability of Agro-Forestry Ecosystems. In Microbial Probiotics for Agricultural Systems: Advances in Agronomic Use; Zúñiga-Dávila, D., González-Andrés, F., Ormeño-Orrillo, E., Eds.; Sustainability in Plant and Crop Protection; Springer International Publishing: Cham, Switzerland, 2019; pp. 71–82. ISBN 978-3-030-17597-9. [Google Scholar]
- Pandya, M.; Rajput, M.; Rajkumar, S. Exploring plant growth promoting potential of non rhizobial root nodules endophytes of Vigna radiata. Microbiology 2015, 84, 80–89. [Google Scholar] [CrossRef]
- Saini, R.; Dudeja, S.S.; Giri, R.; Kumar, V. Isolation, characterization, and evaluation of bacterial root and nodule endophytes from chickpea cultivated in Northern India. J. Basic Microbiol. 2015, 55, 74–81. [Google Scholar] [CrossRef]
- Zhao, L.; Xu, Y.; Lai, X. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz. J. Microbiol. 2017, 49, 269–278. [Google Scholar] [CrossRef]
- Cardoso, P.; Alves, A.; Silveira, P.; Sá, C.; Fidalgo, C.; Freitas, R.; Figueira, E. Bacteria from nodules of wild legume species: Phylogenetic diversity, plant growth promotion abilities and osmotolerance. Sci. Total Environ. 2018, 645, 1094–1102. [Google Scholar] [CrossRef]
- Ampomah, O.Y.; Huss-Danell, K. Genetic diversity of root nodule bacteria nodulating Lotus corniculatus and Anthyllis vulneraria in Sweden. Syst. Appl. Microbiol. 2011, 34, 267–275. [Google Scholar] [CrossRef]
- Sánchez, M.; Ramírez-Bahena, M.-H.; Peix, A.; Lorite, M.J.; Sanjuán, J.; Velázquez, E.; Monza, J. Phyllobacterium loti sp. nov. isolated from nodules of Lotus corniculatus. Int. J. Syst. Evol. Microbiol. 2014, 64, 781–786. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).