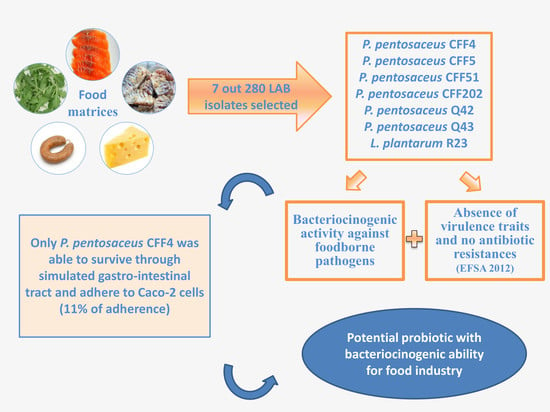
Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
2.2. Screening of Antimicrobial Activity of each LAB Isolate
2.3. LAB Identification
2.3.1. 16S rRNA Sequencing
2.3.2. Whole-Genome Sequencing
2.4. Safety Criteria of Potential Probiotics
2.4.1. Presence of Virulence Factors
Screening-Test of Biogenic Amines Production
Production of Hydrolytic Enzymes
Presence of Virulence Genes
2.4.2. Antibiotic Resistance
2.5. Functional Criteria of Potential Probiotics
2.5.1. Inoculum
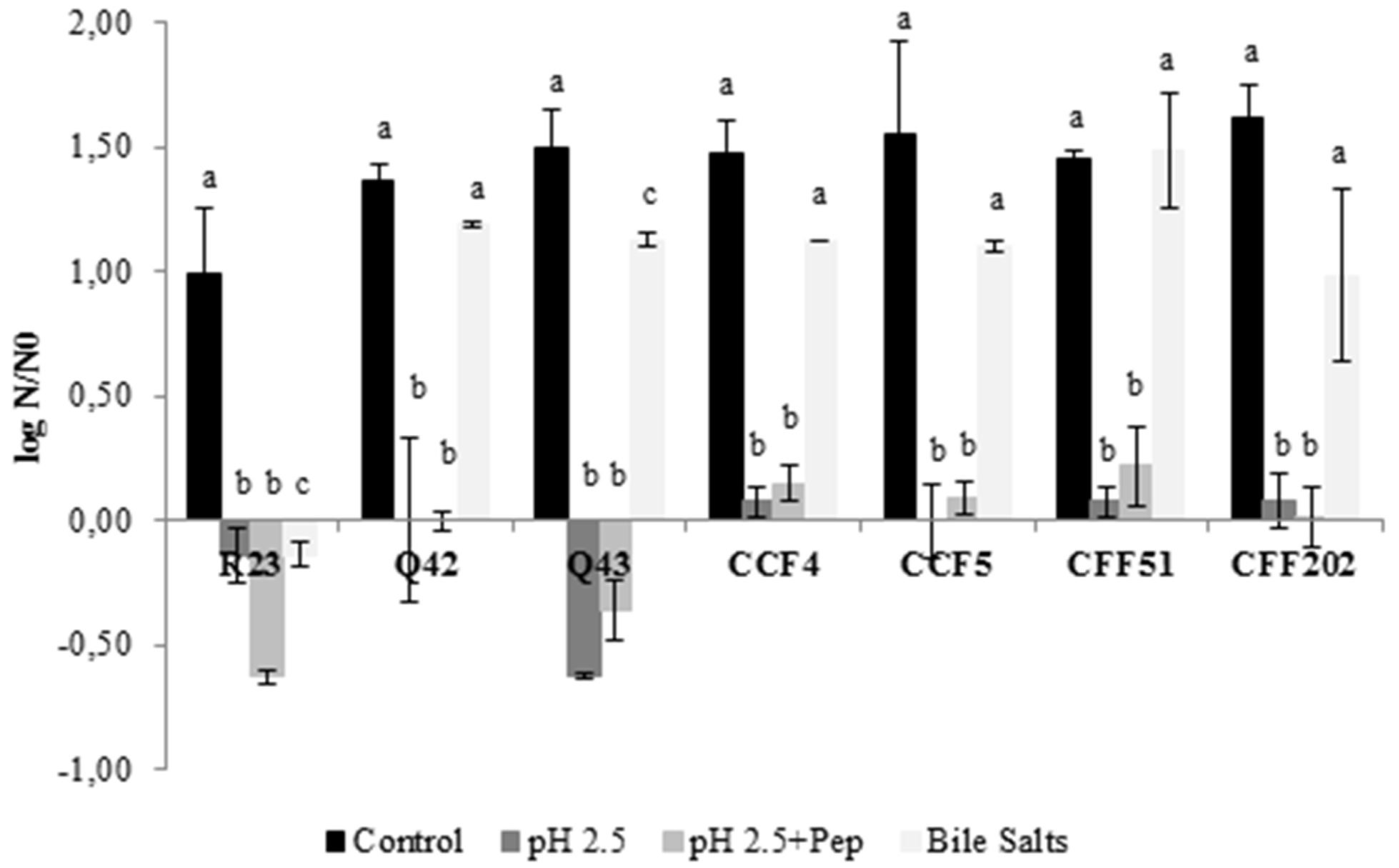
2.5.2. Ability to Resist pH 2.5, pH 2.5 with Pepsin and Bile Salts
2.5.3. Resistance to Simulated Gastrointestinal Tract Conditions
2.5.4. Ability to Adhere to Human Colon Adenocarcinoma Cell Lines Caco-2
Preparation of Cell Lines Caco-2
Preparation of LAB
In Vitro Adherence Assays
2.6. Physiological Criteria of Potential Probiotics
Screening of the Isolates for Bile-Salt Hydrolase (BSH) Activity
2.7. Statistical Analysis
3. Results and Discussion
3.1. Screening of Antimicrobial Activity of each LAB Isolate and Its Identification by 16S rRNA Sequencing
3.2. In Vitro Screening of Probiotic Properties of Selected LAB
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Food and Agriculture Organization/World Health Organization. “Guidelines for the Evaluation of Probiotics in Foods”, Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; World Health Organization: Geneva, Switzerland, 2002. [Google Scholar]
- Amara, A.A.; Shibl, A. Role of probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm. J. 2015, 23, 107–114. [Google Scholar] [CrossRef]
- Sathyabama, S.; Vijayabharathi, R.; Devi, P.B.; Kumar, M.R.; Priyadarisini, V.B. Screening for probiotic properties of strains isolated from feces of various human groups. J. Microbiol. 2012, 50, 603–612. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Kim, S.-K.; Guevarra, R.B.; Kim, Y.-T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.-H. Role of probiotics in human gut microbiome-associated diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef]
- Jia, F.-F.; Zhang, L.-J.; Pang, X.-H.; Gu, X.-X.; Abdelazez, A.; Liang, Y.; Sun, S.-R.; Meng, X.-C. Complete genome sequence of bacteriocin-producing Lactobacillus plantarum KLDS1. 0391, a probiotic strain with gastrointestinal tract resistance and adhesion to the intestinal epithelial cells. Genomics 2017, 109, 432–437. [Google Scholar] [CrossRef]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.; et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174, 388–1405.e21. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Latta, M.; Ma, W.; Wu, Z.; Chen, P. Probiotics database: A potential source of fermented foods. Int. J. Food Prop. 2019, 22, 198–217. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.; Merrifield, C.A.; Hutkins, R. Probiotics for human use. Nut. Bull. 2018, 43, 212–225. [Google Scholar] [CrossRef]
- World Gastroenterology Organisation Global Guidelines. Probiotics and prebiotics. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=4&ved=2ahUKEwiqjdKgyZLoAhWTOnAKHdO5AdMQFjADegQIARAB&url=https%3A%2F%2Fwww.worldgastroenterology.org%2FUserFiles%2Ffile%2Fguidelines%2Fprobiotics-and-prebiotics-english-2017.pdf&usg=AOvVaw3DKlMGBkSgn_QTY7o_5VAX (accessed on 20 January 2020).
- Zommiti, M.; Bouffartigues, E.; Maillot, O.; Barreau, M.; Szunerits, S.; Sebei, K.; Feuilloley, M.; Connil, N.; Ferchichi, M. In vitro assessment of the probiotic properties and bacteriocinogenic potential of Pediococcus pentosaceus MZF16 isolated from artisanal Tunisian meat “dried ossban”. Front. Microbiol. 2018, 9, 2607. [Google Scholar] [CrossRef]
- Barbosa, J.; Borges, S.; Teixeira, P. Pediococcus acidilactici as a potential probiotic to be used in food industry. Int. J. Food Sci. Technol. 2015, 50, 1151–1157. [Google Scholar] [CrossRef]
- Todorov, S.D.; Holzapfel, W.; Nero, L.A. In vitro evaluation of beneficial properties of bacteriocinogenic Lactobacillus plantarum ST8Sh. Probiotics Antimicrob. Proteins 2016, 9, 194–203. [Google Scholar] [CrossRef]
- Moreno, I.; Marasca, E.T.G.; de Sá, P.B.Z.R.; de Souza Moitinho, J.; Marquezini, M.G.; Alves, M.R.C.; Bromberg, R. Evaluation of probiotic potential of bacteriocinogenic lactic acid bacteria strains isolated from meat products. Probiotics Antimicrob. Proteins 2018, 10, 762–774. [Google Scholar] [CrossRef]
- Barbosa, J.; Borges, S.; Teixeira, P. Selection of potential probiotic Enterococcus faecium isolated from Portuguese fermented food. Int. J. Food Microbiol. 2014, 191, 144–148. [Google Scholar] [CrossRef]
- McLauchlin, J.; Hampton, M.D.; Shah, S.; Threlfall, E.J.; Wieneke, A.A.; Curtis, G.D.W. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 1997, 83, 381–388. [Google Scholar] [CrossRef]
- Albano, H.; Pinho, C.; Leite, D.; Barbosa, J.; Silva, J.; Carneiro, L.; Magalhães, R.; Hogg, T.; Teixeira, P. Evaluation of a bacteriocin-producing strain of Pediococcus acidilactici as a biopreservative for ‘‘Alheira”, a fermented meat sausage. Food Control 2009, 20, 764–770. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Faria, C.; Lopes, A.R.; Svensson, L.; Falsen, E.; Moore, E.R.; Ferreira, A.C.; Nunes, O.C.; Manaia, C.M. Sphingobium vermicomposti sp. nov., isolated from vermicompost. Int. J. Syst. Evol. Microbiol. 2009, 59, 3145–3149. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Shaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening produce for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Tiago, I.; Teixeira, I.; Silva, S.; Chung, P.; Verıssimo, A.; Manaia, C.M. Metabolic and genetic diversity of mesophilic and thermophilic bacteria isolated from composted municipal sludge on poly-e-caprolactones. Curr. Microbiol. 2004, 49, 407–414. [Google Scholar] [CrossRef]
- Ben Omar, N.; Castro, A.; Lucas, R.; Abriouel, H.; Yousif, N.M.K.; Franz, C.M.A.P.; Holzapfel, W.H.; Rubén, P.-P.; Martínez-Canãmero, M.; Gálvez, A. Functional and safety aspects of enterococci isolated from different Spanish foods. Syst. Appl. Microbiol. 2004, 27, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.; Gibbs, P.A.; Teixeira, P. Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control 2010, 21, 651–656. [Google Scholar] [CrossRef]
- European Food Safety Authority. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance by EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). EFSA J. 2012, 10, 2740–2749. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement; CLSI=NCCLS M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Miles, A.A.; Misra, S.S. The estimation of the bactericidal power of blood. J. Hyg. 1938, 38, 732–749. [Google Scholar] [CrossRef]
- Barbosa, J.; Borges, S.; Magalhães, R.; Ferreira, V.; Santos, I.; Silva, J.; Almeida, G.; Gibbs, P.; Teixeira, P. Behaviour of Listeria monocytogenes isolates through gastro-intestinal tract passage simulation, before and after two sub-lethal stresses. Food Microbiol. 2012, 30, 24–28. [Google Scholar] [CrossRef]
- Guglielmotti, D.M.; Marcó, M.B.; Golowczyc, M.; Reinheimer, J.A.; del Quiberoni, A.A.L. Probiotic potential of Lactobacillus delbrueckii strains and their phage resistant mutants. Int. Dairy J. 2007, 17, 916–925. [Google Scholar] [CrossRef]
- Tsai, C.; Lin, P.; Hsieh, Y.; Zhang, Z.Y.; Wu, H.C.; Huang, C.C. Cholesterol-lowering potentials of lactic acid bacteria based on bile-salt hydrolase activity and effect of potent strains on cholesterol metabolism in vitro and in vivo. Sci. World J. 2014, 14, 690752. [Google Scholar] [CrossRef]
- Peng, C.; Borges, S.; Magalhães, R.; Carvalheira, A.; Ferreira, V.; Casquete, R.; Teixeira, P. Characterization of anti-listerial bacteriocin produced by lactic acid bacteria isolated from traditional fermented foods from Cambodia. Int. Food Res. J. 2017, 24, 386–393. [Google Scholar]
- Collins, M.D.; Rodrigues, U.M.; Ash, C.; Aguirre, M.; Farrow, J.A.E.; Martinez-Murcia, A.; Phillips, B.A.; Williams, A.M.; Wallbanks, S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett. 1991, 77, 5–12. [Google Scholar] [CrossRef]
- Albano, H.; Todorov, S.D.; van Reenen, C.A.; Hogg, T.; Dicks, L.M.; Teixeira, P. Characterization of two bacteriocins produced by Pediococcus acidilactici isolated from “Alheira”, a fermented sausage traditionally produced in Portugal. Int. J. Food Microbiol. 2007, 116, 239–247. [Google Scholar] [CrossRef]
- Sadishkumar, V.; Kolanchiammal, R.; Jeevaratnam, K. The effect of Piper betle leaves on lacto-fermentation of idli batter, characterization and applicability of potent isolates in soymilk fermentation. Food Sci. Biotechnol. 2015, 24, 2137–2144. [Google Scholar] [CrossRef]
- Abushelaibi, A.; Al-Mahadin, S.; El-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT—Food Sci. Technol. 2017, 79, 316–325. [Google Scholar] [CrossRef]
- Hernández-Alcántara, A.M.; Wacher, C.; Llamas, M.G.; López, P.; Pérez-Chabela, M.L. Probiotic properties and stress response of thermotolerant lactic acid bacteria isolated from cooked meat products. LWT—Food Sci. Technol. 2018, 91, 249–257. [Google Scholar] [CrossRef]
- Mulaw, G.; Tessema, T.S.; Muleta, D.; Tesfaye, A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int. J. Microbiol. 2019, 2019, 7179514. [Google Scholar] [CrossRef]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013, 13, 15–36. [Google Scholar] [CrossRef]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with functional properties: An approach to increase safety and shelf-life of fermented foods. BioMed. Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef]
- Rozman, V.; Lorbeg, P.M.; Accetto, T.; Matijašić, B.B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food Microbiol. 2020, 314, 108388. [Google Scholar] [CrossRef]
- European Food Safety Authority. Opinion of the Scientific Committee on a request from EFSA related to a generic approach to the safety assessment by EFSA of microorganisms used in food/feed and the production of food/feed additives. EFSA J. 2005, 226, 1–12. [Google Scholar]
- European Food Safety Authority. Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 2005, 223, 1–12. [Google Scholar]
- Vera-Pingitore, E.; Jimenez, M.; Dallagnol, A.; Belfiore, C.; Fontana, C.; Fontana, P.; von Wright, A.; Vignolo, G.; Plumed-Ferrer, C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT—Food Sci. Technol. 2016, 71, 288–294. [Google Scholar] [CrossRef]
- García-Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.; Nicoli, J.; Moreira-Silva, J.; Rodríguez, Z.; Fuertes, H.; Nuñez, O.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 2016, 108, 125–132. [Google Scholar] [CrossRef]
- Damodharan, K.; Lee, Y.S.; Palaniyandi, S.A.; Yang, S.H.; Suh, J.-W. Preliminary probiotic and technological characterization of Pediococcus pentosaceus strain KID7 and in vivo assessment of its cholesterol-lowering activity. Front. Microbiol. 2015, 6, 768. [Google Scholar] [CrossRef]
- Han, Q.; Kong, B.; Chen, Q.; Sun, F.; Zhang, H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J. Funct. Foods 2017, 32, 391–400. [Google Scholar] [CrossRef]
- Duary, R.K.; Rajput, Y.S.; Batish, V.K.; Grover, S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J. Med. Res. 2011, 134, 664–671. [Google Scholar]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile Salt Hydrolase Activity in Probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Noriega, L.; Cuevas, I.; Margolles, A.; de los Reyes-Gavilán, C.G. Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int. Dairy J. 2006, 16, 850–855. [Google Scholar] [CrossRef]
| Microorganisms | Species | Source |
|---|---|---|
| Gram-positive | Bacillus cereus Bacillus subtilis Bacillus stearothermophilus Listeria innocua 2030c Staphylococcus aureus 18N (methicillin-resistant > Staphylococcus aureus-MRSA) Staphylococcus aureus 2037 M1 (methicillin- sensitive Staphylococcus aureus-MSSA) | ESB culture collection |
| Enterococcus faecalis ATCC 29212 Staphylococcus aureus ATCC 29213 | ATCC | |
| Enterococcus faecalis DSMZ 12956 Enterococcus faecium DSMZ 13590 Enterococcus flavescens DSMZ 7370 Enterococcus casseliflavus DSMZ 20680 Enterococcus gallinarum DSMZ 20628 | DSMZ | |
| Listeria monocytogenes L7946 Listeria monocytogenes L7947 | McLauchlin et al. [16] | |
| Gram-negative | Acinetobacter baumannii R Acinetobacter baumannii S-1 Acinetobacter baumannii S-2 Acinetobacter calcoaceticus R Acinetobacter calcoaceticus S Klebsiella pneumoniae Proteus mirabilis Proteus vulgaris Pseudomonas aeruginosa Salmonella Braenderup Salmonella Enteritidis Salmonella Typhimurium Yersinia enterocolitica | ESB culture collection |
| Escherichia coli ATCC 25922 | ATCC | |
| Yersinia enterocolitica NCTC 10406 | NCTC | |
| Yeasts | Candida albicans Saccharomyces cerevisiae | ESB |
| Amp | Gen | Kan | Str | Ery | Chl | Tet | |
|---|---|---|---|---|---|---|---|
| R23 | 0.5 | ≤4 | ≤16 | n.r. | ≤0,5 | ≤2 | ≤4 |
| Q42 | 1 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| Q43 | 2 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| CFF4 | 2 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| CFF5 | 2 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| CFF51 | 1 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| CFF202 | 1 | ≤4 | ≤16 | ≤32 | ≤0,5 | ≤2 | ≤4 |
| Log CFU/mL (*) | |||
|---|---|---|---|
| Isolate | 0 min | 60 min (#) | 120 min (†) |
| L. plantarum R23 | 6.82 ± 0.06Aa | 4.49 ± 0.04Ba | 4.56 ± 0.05Ba |
| P. pentosaceus Q42 | 7.16 ± 0.02Aa | 6.82 ± 0.06Ab | 6.64 ± 0.09Ab |
| P. pentosaceus Q43 | 6.87 ± 0.20Aa | 7.18 ± 0.20Ab | 6.65 ± 0.09Ab |
| P. pentosaceus CFF4 | 7.25 ± 0.12Aa | 7.12 ± 0.03Ab | 6.90 ± 0.22Abc |
| P. pentosaceus CFF5 | 7.25 ± 0.03Aa | 7.02 ± 0.17Ab | 7.17 ± 0.13Ac |
| P. pentosaceus CFF51 | 7.06 ± 0.27Aa | 7.30 ± 0.19Ab | 7.04 ± 0.05Abc |
| P. pentosaceus CFF202 | 7.12 ± 0.07Aa | 7.07 ± 0.16Ab | 6.87 ± 0.06Abc |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, A.; Barbosa, J.; Albano, H.; Isidro, J.; Teixeira, P. Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics. Microorganisms 2020, 8, 393. https://doi.org/10.3390/microorganisms8030393
Pinto A, Barbosa J, Albano H, Isidro J, Teixeira P. Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics. Microorganisms. 2020; 8(3):393. https://doi.org/10.3390/microorganisms8030393
Chicago/Turabian StylePinto, Ana, Joana Barbosa, Helena Albano, Joana Isidro, and Paula Teixeira. 2020. "Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics" Microorganisms 8, no. 3: 393. https://doi.org/10.3390/microorganisms8030393
APA StylePinto, A., Barbosa, J., Albano, H., Isidro, J., & Teixeira, P. (2020). Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics. Microorganisms, 8(3), 393. https://doi.org/10.3390/microorganisms8030393