Identification of Species and Subspecies of Lactic Acid Bacteria Present in Spanish Cheeses Type “Torta” by MALDI-TOF MS and pheS gene Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains Isolation
2.2. MALDI-TOF MS Performing and Data Analysis
2.3. Phylogenetic Analysis of pheS Gene
2.4. Genome Analysis of the Subspecies from the Species Identified in this Study
3. Results
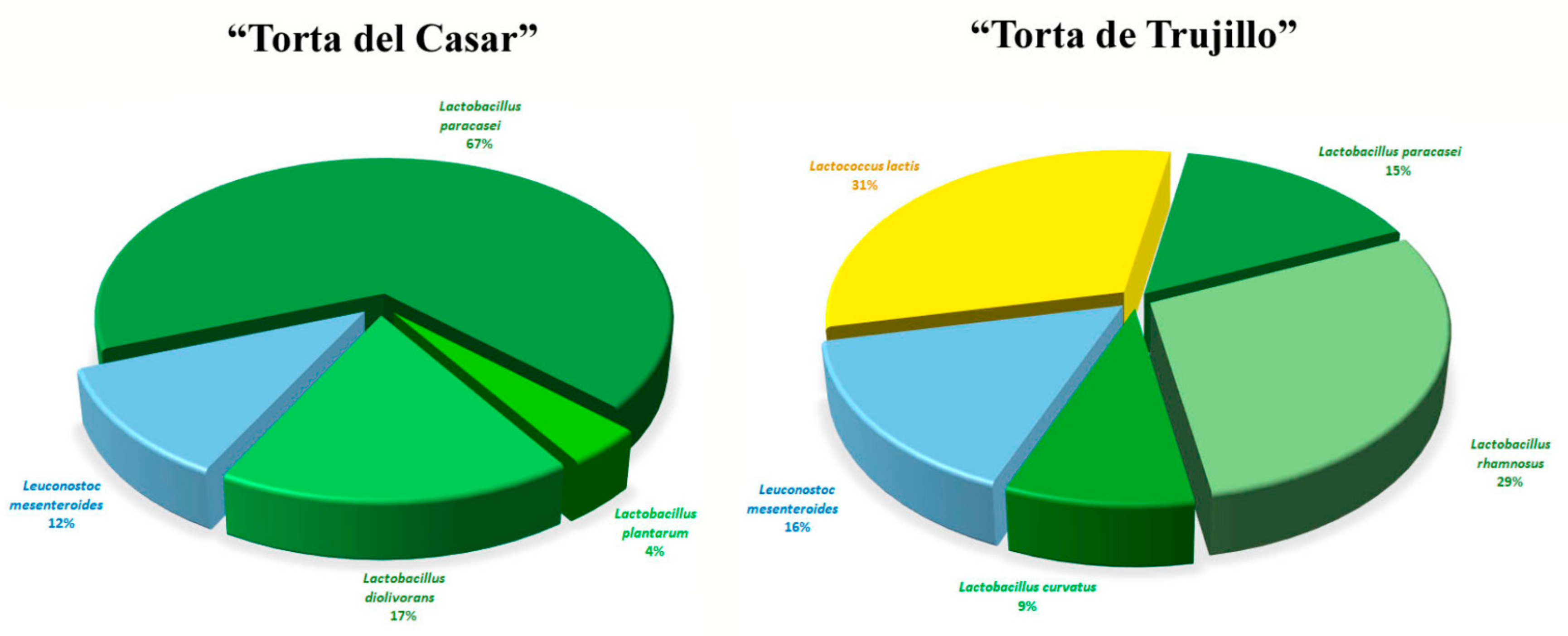
3.1. MALDI-TOF MS Analysis
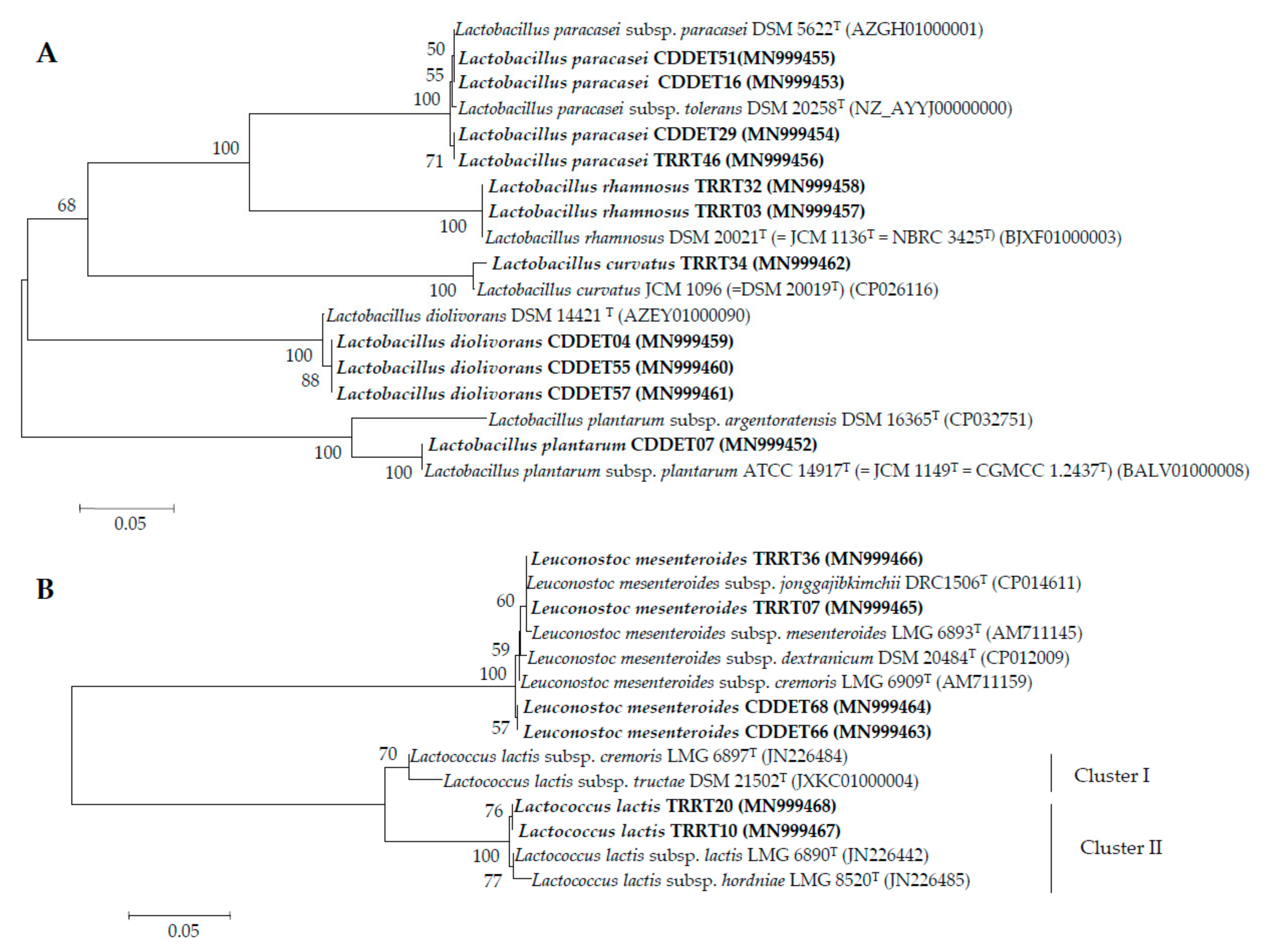
3.2. pheS Gene Analysis
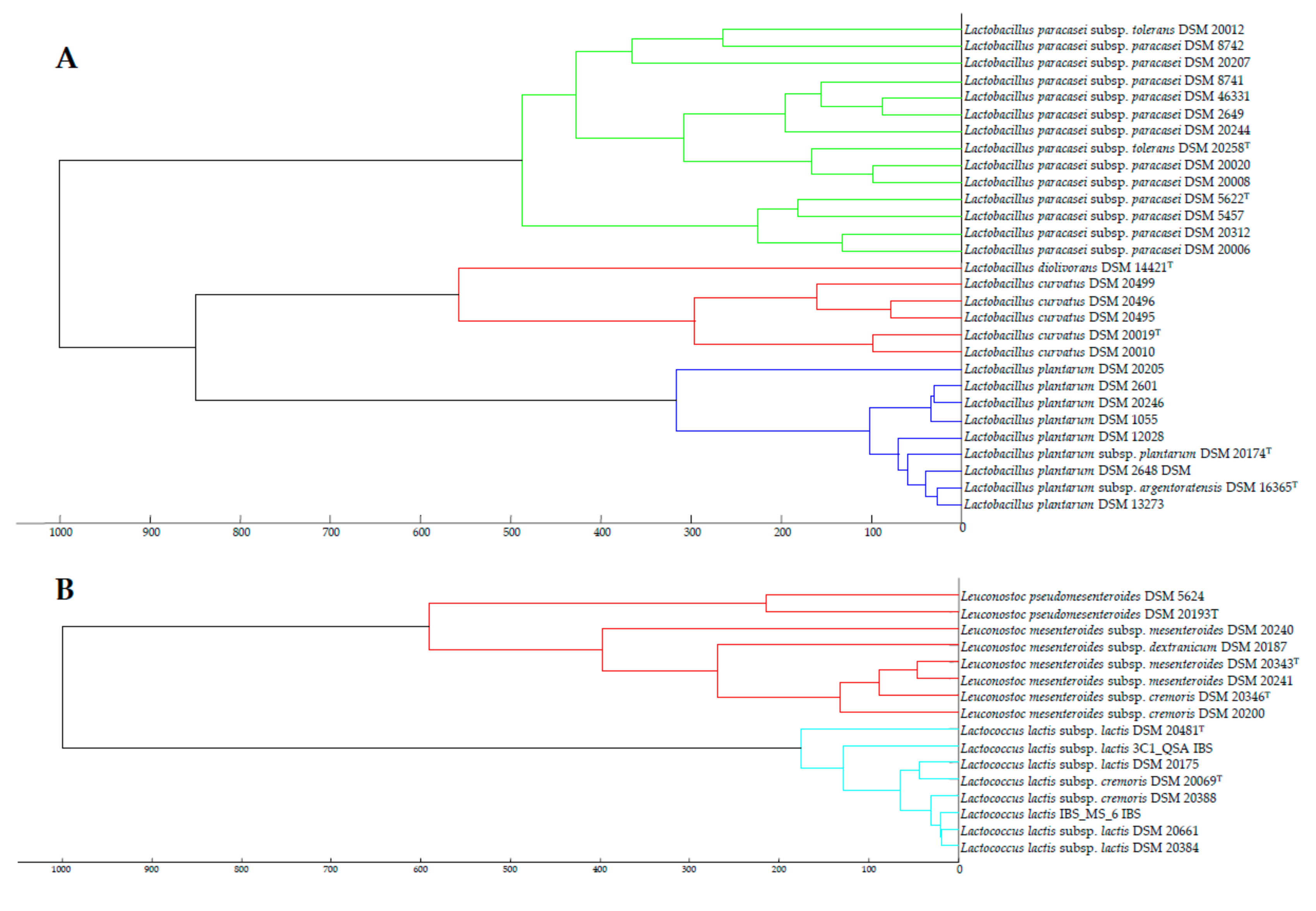
3.3. Taxonomic Status of the Subspecies from the Species Identified in this Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LAB | Lactic acid bacteria |
| La. | Lactobacillus |
| Le. | Leuconostoc |
| Lc. | Lactococcus |
References
- Ludwig, W.; Schleifer, K.H.; Whitman, W.B. Lactobacillales ord. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Zielińska, D.; Kolożyn-Krajewska, D. Food-origin lactic acid bacteria may exhibit probiotic properties: Review. Biomed Res. Int. 2018, 2018, 5063185. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, L.; Bleckwedel, J.; Ortiz, E.; Pescuma, M.; Mozzi, F. Lactic Acid Bacteria. In Industrial Biotechnology; Wittmann, C., Liao, J.C., Eds.; Wiley-VCH: Weinheim, Germany, 2016; pp. 395–451. [Google Scholar]
- Poullet, B.; Huertas, M.; Sánchez, A.; Cáceres, P.; Larriba, G. Main lactic acid bacteria isolated during ripening of Casar de Caceres cheese. J. Dairy Res. 1993, 60, 123–127. [Google Scholar] [CrossRef]
- Ordiales, J.; Benito, M.J.; Martín, A.; Casquete, R.; Serradilla, M.J.; de Guía Córdoba, M. Bacterial communities of the traditional raw ewe’s milk cheese “Torta del Casar” made without the addition of a starter. Food Control 2013, 33, 448–454. [Google Scholar] [CrossRef]
- Morandi, S.; Brasca, M.; Lodi, R. Technological, phenotypic and genotypic characterisation of wild lactic acid bacteria involved in the production of Bitto PDO Italian cheese. Dairy Sci. Technol. 2011, 91, 341–359. [Google Scholar] [CrossRef]
- Colombo, E.; Franzetti, L.; Frusca, M.; Scarpellini, M. Phenotypic and genotypic characterization of lactic acid bacteria isolated from Artisanal Italian goat cheese. J. Food Prot. 2010, 73, 657–662. [Google Scholar] [CrossRef]
- Pogačić, T.; Mancini, A.; Santarelli, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Gatti, M. Diversity and dynamic of lactic acid bacteria strains during aging of a long ripened hard cheese produced from raw milk and undefined natural starter. Food Microbiol. 2013, 36, 207–215. [Google Scholar] [CrossRef]
- Pangallo, D.; Saková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, L.; Kuchta, T. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, R.; Tuohy, K.M. Biodiversity and γ-aminobutyric acid production by lactic acid bacteria isolated from traditional alpine raw cow’s milk cheeses. Biomed Res. Int. 2015, 2015, 625740. [Google Scholar] [CrossRef]
- Domingos-Lopes, M.F.P.; Stanton, C.; Ross, P.R.; Dapkevicius, M.L.E.; Silva, C.C.G. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017, 63, 178–190. [Google Scholar] [CrossRef]
- Foschi, C.; Laghi, L.; Parolin, C.; Giordani, B.; Compri, M.; Cevenini, R.; Marangoni, A.; Vitali, B. Novel approaches for the taxonomic and metabolic characterization of lactobacilli: Integration of 16S rRNA gene sequencing with MALDI-TOF MS and 1H-NMR. PLoS ONE 2017, 12, e0172483. [Google Scholar] [CrossRef]
- Nacef, M.; Chevalier, M.; Chollet, S.; Drider, D.; Flahaut, C. MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int. J. Food Microbiol. 2017, 247, 2–8. [Google Scholar] [CrossRef]
- Sato, H.; Torimura, M.; Kitahara, M.; Ohkuma, M.; Hotta, Y.; Tamura, H. Characterization of the Lactobacillus casei group based on the profiling of ribosomal proteins coded in S10-spc-alpha operons as observed by MALDI-TOF MS. Syst. Appl. Microbiol. 2012, 35, 447–454. [Google Scholar] [CrossRef]
- Soro-Yao, A.A.; Schumann, P.; Thonart, P.; Djè, K.M.; Pukall, R. The use of MALDI-TOF Mass Spectrometry, ribotyping and phenotypic tests to identify lactic acid bacteria from fermented cereal foods in Abidjan (Côte d’Ivoire). Open Microbiol. J. 2014, 8, 78–86. [Google Scholar] [CrossRef]
- Tanigawa, K.; Kawabata, H.; Watanabe, K. Identification and typing of Lactococcus lactis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 2010, 76, 4055–4062. [Google Scholar] [CrossRef]
- Doan, N.T.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; Aerts, M.; Le Thanh, B.; Vandamme, P. Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in Northern Vietnam. Lett. Appl. Microbiol. 2012, 55, 265–273. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; Aerts, M.; Binh Thanh, L.; Vandamme, P. A description of the lactic acid bacteria microbiota associated with the production of traditional fermented vegetables in Vietnam. Int. J. Food Microbiol. 2013, 163, 19–27. [Google Scholar] [CrossRef]
- Ferreira, L.; Sánchez-Juanes, F.; García-Fraile, P.; Rivas, R.; Mateos, P.F.; Martínez-Molina, E.; González-Buitrago, J.M.; Velázquez, E. MALDI-TOF mass spectrometry is a fast and reliable platform for identification and ecological studies of species from family Rhizobiaceae. PLoS ONE 2011, 6, e20223. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustalX windows interface: Flexible strategies for multiple sequence alignement aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. A neighbour-joining method: A new method for reconstructing phylogenetics trees. Mol. Biol. Evol. 1987, 44, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 3, 1870–1874. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Mora, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Doijad, S.; Chakraborty, T. Genome-based analyses indicate that Serratia marcescens subsp. marcescens and Serratia marcescens subsp. sakuensis do not merit separation to subspecies status. Int. J. Syst. Evol. Microbiol. 2019, 69, 3924–3926. [Google Scholar]

| Torta del Casar | |||
|---|---|---|---|
| Strains | Closest Taxa | Score Values | Groups |
| CCDET 01 | La. paracasei subsp. paracasei DSM 20006 | 2.502 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.194 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.960 | ||
| CCDET 04 | La. diolivorans DSM 14421T | 2.228 | IIB |
| CCDET 05 | La. paracasei subsp. paracasei DSM 20006 | 2.504 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.193 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.174 | ||
| CCDET 07 | La. plantarum DSM 2601 | 2.478 | IV |
| La. plantarum subsp. argentoratensis DSM 16365T | 2.322 | ||
| La. plantarum subsp. plantarum DSM 20174T | 2.037 | ||
| CCDET 09 | La. paracasei subsp. paracasei DSM 20006 | 2.511 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.128 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.476 | ||
| CCDET 10 | La. paracasei subsp. paracasei DSM 20006 | 2.483 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.097 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.051 | ||
| CCDET 11 | La. paracasei subsp. paracasei DSM 20244 | 2.517 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.063 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.911 | ||
| CCDET 12 | La. paracasei subsp. paracasei DSM 20006 | 2.433 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.113 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.018 | ||
| CCDET 13 | La. diolivorans DSM 14421T | 2.218 | IIB |
| CCDET 14 | La. paracasei subsp. paracasei DSM 2649 | 2.43 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.047 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.773 | ||
| CCDET 15 | La. paracasei subsp. paracasei DSM 20006 | 2.531 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.053 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.542 | ||
| CCDET 16 | La. paracasei subsp. paracasei DSM 20006 | 2.545 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.147 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.112 | ||
| CCDET 18 | La. paracasei subsp. paracasei DSM 20006 | 2.463 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.107 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.911 | ||
| CCDET19 | La. paracasei subsp. paracasei DSM 20244 | 2.500 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.309 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.103 | ||
| CCDET20 | Le. mesenteroides subsp. dextranicum DSM 20187 | 2.062 | IA |
| Le. mesenteroides subsp. mesenteroides DSM 20343T | 1.683 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.648 | ||
| CCDET21 | La. paracasei subsp. paracasei DSM 20006 | 2.337 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.174 | ||
| La. paracasei subsp. paracasei DSM 5622T | 1.952 | ||
| CCDET 22 | La. paracasei subsp. paracasei DSM 20006 | 2.380 | VIIB |
| La. paracasei subsp. paracasei DSM 5622TLa. paracasei subsp. tolerans DSM 20258T | 2.0401.752 | ||
| CCDET 23 | La. paracasei subsp. paracasei DSM 20244 | 2.397 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 1.938 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.800 | ||
| CCDET 24 | La. paracasei subsp. paracasei DSM 20312 | 2.355 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.038 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.897 | ||
| CCDET 25 | La. paracasei subsp. paracasei DSM 20006 | 2.544 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.115 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.092 | ||
| CCDET 26 | La. paracasei subsp. paracasei DSM 20006 | 2.476 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.165 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.033 | ||
| CCDET 27 | La. plantarum subsp. plantarum DSM 12028 | 2.177 | IV |
| La. plantarum subsp. argentoratensis DSM 16365T | 2.131 | ||
| La. plantarum subsp. plantarum DSM 20174T | 1.963 | ||
| CCDET 28 | La. paracasei subsp. paracasei DSM 20244 | 2.386 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.097 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.097 | ||
| CCDET 29 | La. paracasei subsp. paracasei DSM 46331 | 2.432 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.224 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.072 | ||
| CCDET 30 | La. paracasei subsp. paracasei DSM 20006 | 2.492 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.157 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.003 | ||
| CCDET 32 | La. paracasei subsp. paracasei DSM 20006 | 2.513 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.113 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.540 | ||
| CCDET 34 | La. paracasei subsp. paracasei DSM 20244 | 2.444 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.160 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.120 | ||
| CCDET 35 | La. paracasei subsp. paracasei DSM 20006 | 2.475 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.080 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.962 | ||
| CCDET 38 | La. paracasei subsp. paracasei DSM 20006 | 2.452 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.112 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.083 | ||
| CCDET 39 | La. paracasei subsp. paracasei DSM 20006 | 2.494 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.059 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.847 | ||
| CCDET 42 | La. paracasei subsp. paracasei DSM 20006 | 2.223 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 1.871 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.854 | ||
| CCDET 43 | La. paracasei subsp. paracasei DSM 20244 | 2.348 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.035 | ||
| La. paracasei subsp. tolerans DSM 20258T | 1.990 | ||
| CCDET 44 | La. paracasei subsp. paracasei DSM 20244 | 2.339 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.106 | ||
| La. paracasei subsp. paracasei DSM 5622T | 1.998 | ||
| CCDET 45 | La. paracasei subsp. paracasei DSM 20244 | 2.353 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.061 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.053 | ||
| CCDET 46 | La. diolivorans DSM 14421T | 2.235 | IIB |
| CCDET51 | La. paracasei subsp. paracasei DSM 20244 | 2.437 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.054 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.054 | ||
| CCDET52 | Le. mesenteroides subsp. dextranicum DSM 20187 | 2.000 | IA |
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.690 | ||
| Le. mesenteroides subsp. mesenteroides DSM 20343T | 1.374 | ||
| CCDET53 | Le. mesenteroides subsp. mesenteroides DSM 20241 | 2.120 | IA |
| Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.106 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.961 | ||
| CCDET 54 | La. diolivorans DSM 14421T | 2.003 | IIB |
| CCDET 55 | La. diolivorans DSM 14421T | 1.911 | IIB |
| CCDET 56 | La. diolivorans DSM 14421T | 2.093 | IIB |
| CCDET 57 | La. diolivorans DSM 14421T | 2.100 | IIB |
| CCDET 58 | La. diolivorans DSM 14421T | 2.149 | IIB |
| CCDET 59 | La. diolivorans DSM 14421T | 2.106 | IIB |
| CCDET 61 | La. paracasei subsp. paracasei DSM 20006 | 2.353 | VIIA |
| La. paracasei subsp. paracasei DSM 5622T | 2.170 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.101 | ||
| CCDET62 | La. paracasei subsp. paracasei DSM 20244 | 2.536 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.157 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.148 | ||
| CCDET 63 | La. paracasei subsp. paracasei DSM 8741 | 2.383 | VIIA |
| La. paracasei subsp. paracasei DSM 5622T | 2.100 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.082 | ||
| CCDET64 | La. paracasei subsp. paracasei DSM 20244 | 2.493 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.149 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.092 | ||
| CCDET65 | Le. mesenteroides subsp. dextranicum DSM 20187 | 2.072 | IA |
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.692 | ||
| Le. mesenteroides subsp. mesenteroides DSM 20343T | 1.454 | ||
| CCDET66 | Le. mesenteroides subsp. dextranicum DSM 20187 | 2.071 | IA |
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.633 | ||
| Le. mesenteroides subsp. mesenteroides DSM 20343T | 1.355 | ||
| CCDET67 | La. paracasei subsp. paracasei DSM 20244 | 2.468 | VIIA |
| La. paracasei subsp. tolerans DSM 20258T | 2.233 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.047 | ||
| CCDET68 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.204 | IA |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.089 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.953 | ||
| Torta de Trujillo | |||
| Strains | Closest taxa | Score values | Groups |
| TRRT01 | La. rhamnosus CIP A157T | 2.362 | VI |
| TRRT02 | Lc. lactis subsp. lactis DSM 20661 | 2.433 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.149 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.913 | ||
| TRRT03 | La. rhamnosus CIP A157T | 2.389 | VI |
| TRRT04 | La. curvatus DSM 20499 | 2.430 | V |
| La. curvatus DSM 20019T | 2.007 | ||
| TRRT05 | La. paracasei subsp. paracasei DSM 20006 | 2.393 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.193 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.109 | ||
| TRRT06 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.368 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.097 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.963 | ||
| TRRT07 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.380 | IB |
| Le. mesenteroides subsp. cremoris DSM 20346T | 2.221 | ||
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.026 | ||
| TRRT08 | Lc. lactis subsp. lactis DSM 20661 | 2.373 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.209 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.848 | ||
| TRRT09 | Lc. lactis subsp. lactis DSM 20661 | 2.283 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.214 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.896 | ||
| TRRT10 | Lc. lactis subsp. lactis DSM 20661 | 2.236 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.198 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.771 | ||
| TRRT11 | La. rhamnosus CIP A157T | 2.366 | VI |
| TRRT12 | Lc. lactis subsp. lactis DSM 20661 | 2.310 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.150 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.983 | ||
| TRRT13 | Lc. lactis subsp. lactis DSM 20661 | 2.392 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.255 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.988 | ||
| TRRT14 | Lc. lactis subsp. lactis DSM 20661 | 2.371 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.223 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.901 | ||
| TRRT15 | La. rhamnosus CIP A157T | 2.324 | VI |
| TRRT16 | Lc. lactis subsp. lactis DSM 20661 | 2.456 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.228 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.868 | ||
| TRRT17 | La. rhamnosus CIP A157T | 2.360 | VI |
| TRRT18 | Lc. lactis subsp. lactis DSM 20661 | 2.521 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.215 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.998 | ||
| TRRT19 | Lc. lactis subsp. lactis DSM 20661 | 2.514 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.196 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.927 | ||
| TRRT20 | Lc. lactis subsp. lactis DSM 20661 | 2.461 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.157 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.992 | ||
| TRRT21 | Lc. lactis subsp. lactis DSM 20661 | 2.538 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.226 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 2.045 | ||
| TRRT22 | Lc. lactis subsp. lactis DSM 20661 | 2.345 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.286 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 1.955 | ||
| TRRT23 | La. rhamnosus CIP A157T | 2.426 | VI |
| TRRT24 | Lc. lactis subsp. lactis DSM 20661 | 2.468 | III |
| Lc. lactis subsp. lactis DSM 20481T | 2.243 | ||
| Lc. lactis subsp. cremoris DSM 20069T | 2.036 | ||
| TRRT25 | La. paracasei subsp. paracasei DSM 20006 | 2.425 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.181 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.144 | ||
| TRRT26 | La. rhamnosus CIP A157T | 2.357 | VI |
| TRRT28 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.358 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.090 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 2.044 | ||
| TRRT30 | La. curvatus DSM 20499 | 2.340 | V |
| La. curvatus DSM 20019T | 2.116 | ||
| TRRT31 | La. rhamnosus CIP A157T | 2.433 | VI |
| TRRT32 | La. rhamnosus CIP A157T | 2.350 | VI |
| TRRT33 | La. paracasei subsp. paracasei DSM 20006 | 2.435 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.136 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.103 | ||
| TRRT34 | La. curvatus DSM 20499 | 2.405 | V |
| La. curvatus DSM 20019T | 2.166 | ||
| TRRT35 | La. rhamnosus CIP A157T | 2.367 | VI |
| TRRT36 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.389 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.035 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.985 | ||
| TRRT37 | La. paracasei subsp. paracasei DSM 20006 | 2.448 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.114 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.000 | ||
| TRRT38 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.359 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.131 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.908 | ||
| TRRT39 | La. curvatus DSM 20499 | 2.234 | V |
| La. curvatus DSM 20019T | 2.118 | ||
| TRRT40 | La. rhamnosus CIP A157T | 2.361 | VI |
| TRRT41 | La. rhamnosus CIP A157T | 2.354 | VI |
| TRRT42 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.308 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.004 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.982 | ||
| TRRT43 | La. paracasei subsp. paracasei DSM 20006 | 2.362 | VIIB |
| La. paracasei subsp. paracasei DSM 5622T | 2.234 | ||
| La. paracasei subsp. tolerans DSM 20258T | 2.182 | ||
| TRRT44 | La. rhamnosus CIP A157T | 2.405 | VI |
| TRRT46 | La. paracasei subsp. paracasei DSM 20006 | 2.459 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 2.221 | ||
| La. paracasei subsp. paracasei DSM 5622T | 2.000 | ||
| TRRT47 | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.322 | IB |
| Le. mesenteroides subsp. dextranicum DSM 20187 | 2.041 | ||
| Le. mesenteroides subsp. cremoris DSM 20346T | 1.864 | ||
| TRRT48 | La. paracasei subsp. paracasei DSM 20006 | 2.242 | VIIB |
| La. paracasei subsp. tolerans DSM 20258T | 1.983 | ||
| La. paracasei subsp. paracasei DSM 5622T | 1.658 | ||
| Strains | Closest Species | Score Values MALDI-TOF | pheS Gene Similarity (%) | ANIb (%) | dDDH (%) |
|---|---|---|---|---|---|
| La. plantarum subsp plantarum ATCC 14917T (DSM 20174T) | La. plantarum subsp. argentoratensis DSM 16365T | 2.424 | 90.5% | 94.9 | 62.9 |
| La. paracasei subsp paracasei DSM 5622T | La. paracasei subsp. tolerans DSM 20258T | 1.846 | 99.5 | 97.9 | 84.9 |
| Le. mesenteroides subsp mesenteroides ATCC 8293T (DSM 20343T) | Le. mesenteroides subsp. cremoris ATCC 19254T (DSM 20346T) | 2.456 | 99.5 | 98.1 | 90.9 |
| Le. mesenteroides subsp mesenteroides ATCC 8293T (DSM 20343T) | Le. mesenteroides subsp. dextranicum DSM 20484T | nd | 99.2 | 98.2 | 91.9 |
| Le. mesenteroides subsp mesenteroides ATCC 8293T (DSM 20343T) | Le. mesenteroides subsp. jonggajibkimchii DRC1506T | nd | 99.7 | 98.4 | 90.1 |
| Le. mesenteroides subsp. cremoris ATCC 19254T (DSM 20346T) | Le. mesenteroides subsp. dextranicum DSM 20484T | nd | 99.7 | 98.5 | 91.5 |
| Le. mesenteroides subsp. cremoris ATCC 19254T (DSM 20346T) | Le. mesenteroides subsp. jonggajibkimchii DRC1506T | nd | 99.7 | 98.1 | 88.5 |
| Le. mesenteroides subsp. dextranicum DSM 20484T | Le. mesenteroides subsp. jonggajibkimchii DRC1506T | nd | 99.5 | 98.4 | 90.1 |
| Lc.lactis subsp lactis ATCC 19435T (DSM 20481T) | Lc. lactis subsp. cremoris NBRC 100676T (DSM 20069T) | 2.174 | 92.2 | 86.7 | 32.7 |
| Lc.lactis subsp lactis ATCC 19435T (DSM 20481T) | Lc. lactis subsp. hordniae CCUG 32210T | nd | 99.2 | 96.7 | 79.9 |
| Lc.lactis subsp lactis ATCC 19435T (DSM 20481T) | Lc. lactis subsp. tructae DSM 21502T | nd | 92.5 | 86.1 | 31.7 |
| Lc. lactis subsp. cremoris NBRC 100676T (DSM 20069T) | Lc. lactis subsp. hordniae CCUG 32210T | nd | 91.5 | 86.0 | 31.4 |
| Lc. lactis subsp. cremoris NBRC 100676T (DSM 20069T) | Lc. lactis subsp. tructae DSM 21502T | nd | 98.5 | 97.5 | 83.6 |
| Lc. lactis subsp. hordniae CCUG 32210T | Lc. lactis subsp. tructae DSM 21502T | nd | 91.8 | 85.9 | 31.6 |
| MALDI-TOF MS Group | Number of Strains | Selected Strains | Closest Taxa | Score Values | pheS Gene Similarity (%) |
|---|---|---|---|---|---|
| Group IA | 6 from “Torta del Casar” | CCDET66, | Le. mesenteroides subsp. mesenteroides DSM 20343T | 1.3–2.2 | 99.2 |
| 1 from “Torta de Trujillo” | CCDET68 | Le. mesenteroides subsp. cremoris DSM 20346T | 1.6–2.0 | 99.7 | |
| Group IB * | 6 from “Torta de Trujillo” | TRRT07, | Le. mesenteroides subsp. mesenteroides DSM 20343T | 2.3–2.4 | 99.7 |
| TRRT36 | Le. mesenteroides subsp. cremoris DSM 20346T | 1.8–2.2 | 99.5 | ||
| Branch IIA | 1 from “Torta del Casar” | CCDET55 | La. diolivorans DSM 14421T | 1.9 | 99.3 |
| Group IIB | 9 from “Torta del Casar” | CCDET04, CCDET57 | La. diolivorans DSM 14421T | 1.9–2.2 | 99.3 |
| Group III | 13 from “Torta de Trujillo” | TRRT10, | Lc. lactis subsp. lactis DSM 20481T | 1.9–2.3 | 99.5 |
| TRRT20 | Lc. lactis subsp. cremoris DSM 20069T | 1.7–2.1 | 99.0 | ||
| Group IV | 2 from “Torta del Casar” | CCDET07 | La. plantarum subsp. plantarum DSM 20174T | 2.1–2.3 | 100 |
| La. plantarum subsp. argentoratensis DSM 16365T | 1.9–2.0 | 90.8 | |||
| Group V | 4 from “Torta de Trujillo” | TRRT34 | La. curvatus DSM 20019T | 2.0–2.2 | 99.2 |
| Group VI | 13 from “Torta de Trujillo” | TRRT03, TRRT32 | La. rhamnosus CIP A157T | 2.3–2.4 | 100 |
| VIIA | 8 from “Torta del Casar” | CCDET29 | La. paracasei subsp. paracasei DSM 5622T | 2.0–2.1 | 99.5 |
| La. paracasei subsp. tolerans DSM 20258T | 2.0–2.3 | 99.5 | |||
| VIIA | 8 from “Torta del Casar” | CCDET51 | La. paracasei subsp. paracasei DSM 5622T | 2.0–2.1 | 100 |
| La. paracasei subsp. tolerans DSM 20258T | 2.0–2.3 | 99.7 | |||
| VIIB | 22 from “Torta del Casar” | CCDET16 | La. paracasei subsp. paracasei DSM 5622T | 1.8–2.3 | 100 |
| 7 from “Torta de Trujillo” | |||||
| La. paracasei subsp. tolerans DSM 20258T | 1.9–2.2 | 99.7 | |||
| VIIB | 22 from “Torta del Casar” | TRRT46 | La. paracasei subsp. paracasei DSM 5622T | 1.8-2.3 | 99.5 |
| 7 from “Torta de Trujillo” | La. paracasei subsp. tolerans DSM 20258T | 1.9-2.2 | 99.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Juanes, F.; Teixeira-Martín, V.; González-Buitrago, J.M.; Velázquez, E.; Flores-Félix, J.D. Identification of Species and Subspecies of Lactic Acid Bacteria Present in Spanish Cheeses Type “Torta” by MALDI-TOF MS and pheS gene Analyses. Microorganisms 2020, 8, 301. https://doi.org/10.3390/microorganisms8020301
Sánchez-Juanes F, Teixeira-Martín V, González-Buitrago JM, Velázquez E, Flores-Félix JD. Identification of Species and Subspecies of Lactic Acid Bacteria Present in Spanish Cheeses Type “Torta” by MALDI-TOF MS and pheS gene Analyses. Microorganisms. 2020; 8(2):301. https://doi.org/10.3390/microorganisms8020301
Chicago/Turabian StyleSánchez-Juanes, Fernando, Vanessa Teixeira-Martín, José Manuel González-Buitrago, Encarna Velázquez, and José David Flores-Félix. 2020. "Identification of Species and Subspecies of Lactic Acid Bacteria Present in Spanish Cheeses Type “Torta” by MALDI-TOF MS and pheS gene Analyses" Microorganisms 8, no. 2: 301. https://doi.org/10.3390/microorganisms8020301
APA StyleSánchez-Juanes, F., Teixeira-Martín, V., González-Buitrago, J. M., Velázquez, E., & Flores-Félix, J. D. (2020). Identification of Species and Subspecies of Lactic Acid Bacteria Present in Spanish Cheeses Type “Torta” by MALDI-TOF MS and pheS gene Analyses. Microorganisms, 8(2), 301. https://doi.org/10.3390/microorganisms8020301






