Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Medium
2.2. Sampling and Isolation of Tetracycline (TC)-Degrading Bacteria
2.3. Construction of TC-Degrading Bacterial Consortium
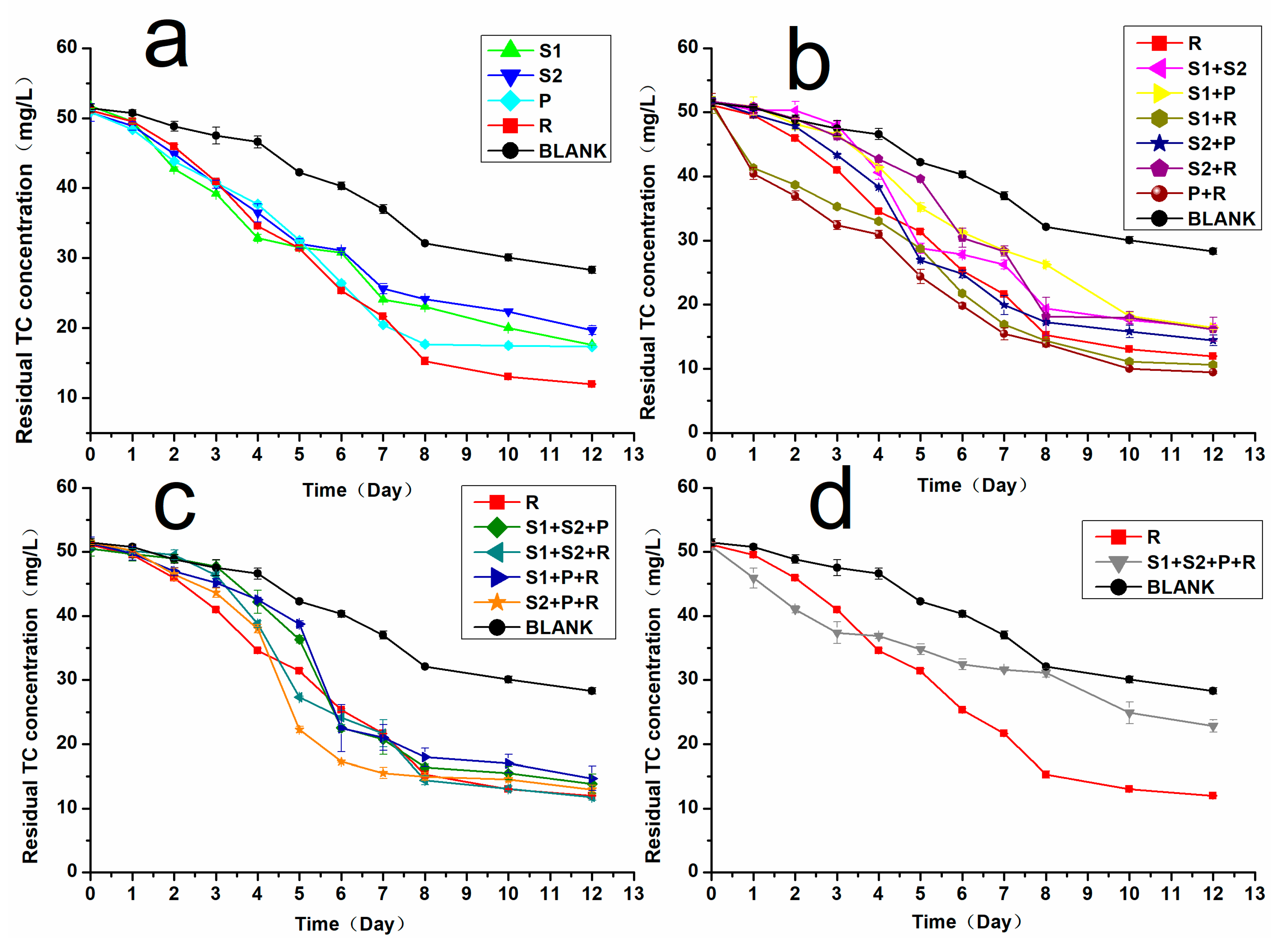
2.4. Lab-Scale Soil Remediation Using the Best TC-Degrading Bacterial Consortium
2.5. Quantification of TC and Its Sample Pretreatment
2.6. DNA Extraction of Soil Samples
2.7. 16S rRNA Gene Amplification, Sequencing, and Data Processing
2.8. Quantification of Tetracycline Resistance Genes (TRGs) and Mobile Gene Elements (MGEs) Using High-Throughput qPCR (HT-qPCR)
3. Results and Discussion
3.1. Strain Isolation and Construction of TC-Degrading Bacterial Consortium
3.2. Effectiveness of the TC-Degrading Bacterial Consortium in the Soil Environment
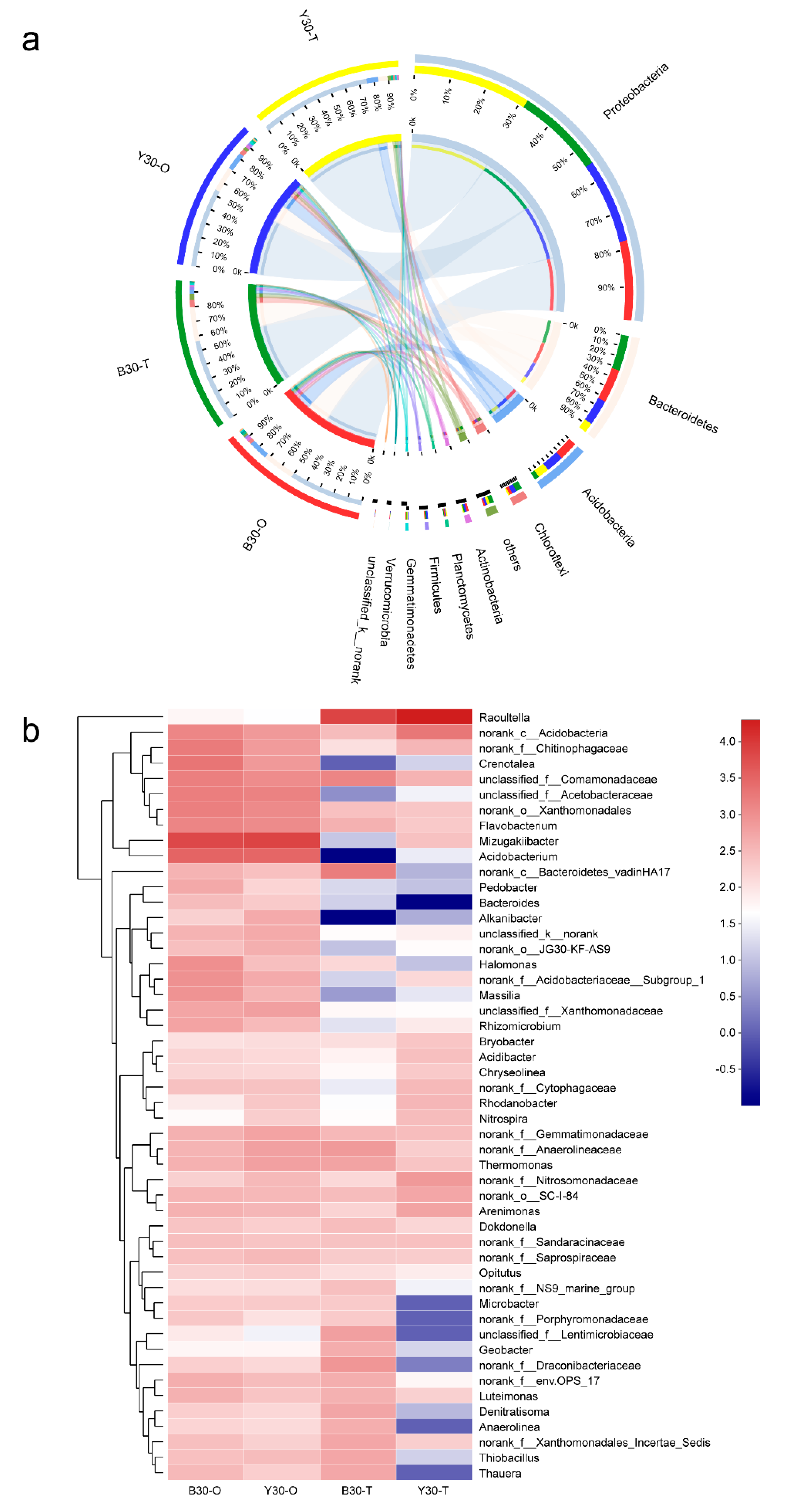
3.3. Shifts of Bacterial Communities during the Lab-Scale Soil Remediation
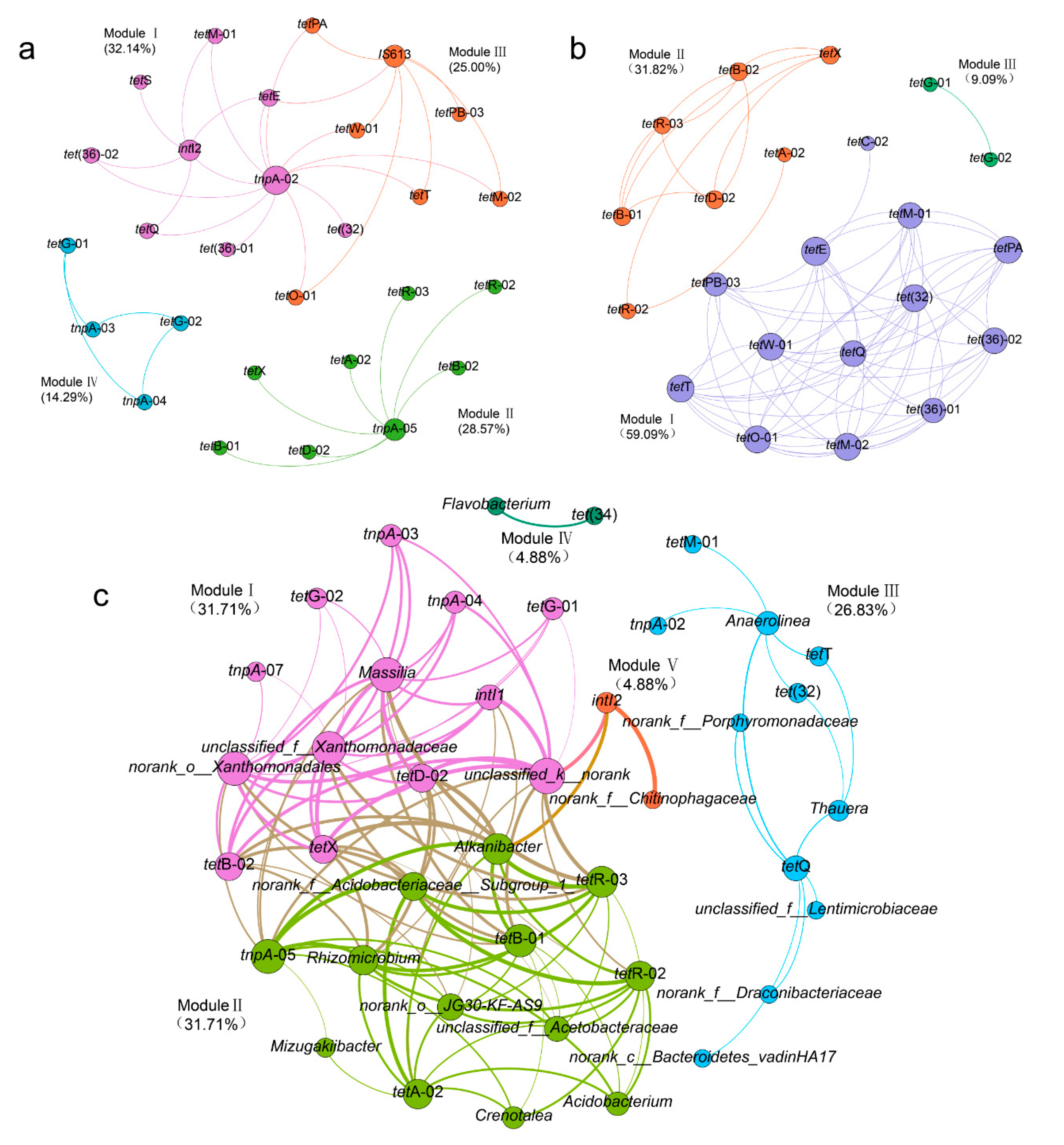
3.4. Variations in TRGs and MGEs during the Lab-Scale Soil Remediation
3.5. Correlations between TRGs, MGEs, and Bacterial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Qiao, M.; Chen, W.; Su, J.; Zhang, B.; Zhang, C. Fate of tetracyclines in swine manure of three selected swine farms in China. J. Environ. Sci. 2012, 24, 1047–1052. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, C.; Lv, J.; Hou, S.; Luo, Y.; Zhang, Y.; Xu, J. Spatiotemporal profile of tetracycline and sulfonamide and their resistance on a catchment scale. Environ. Pollut. 2018, 241, 1098–1105. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Sun, H.; Zhao, L.; Liu, Y. Effect of tetracycline on microbial community structure associated with enhanced biological N&P removal in sequencing batch reactor. Bioresour. Technol. 2018, 256, 414–420. [Google Scholar] [CrossRef]
- Wang, Q.; Li, X.; Yang, Q.; Chen, Y.; Du, B. Evolution of microbial community and drug resistance during enrichment of tetracycline-degrading bacteria. Ecotoxicol. Environ. Saf. 2019, 171, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Wang, X.; Gu, J.; Zhang, S.; Yin, Y.; Li, Y.; Qian, X.; Sun, W. Effects of different swine manure to wheat straw ratios on antibiotic resistance genes and the microbial community structure during anaerobic digestion. Bioresour. Technol. 2017, 231, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Li, Q.; Mei, L.; Zhao, H.; Bai, Y.; Shen, M.; Hu, J. Tetracycline resistance genes are more prevalent in wet soils than in dry soils. Ecotoxicol. Environ. Saf. 2018, 156, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, M.K.; Liu, T.; Chen, H.; Verma, S.; Duan, Y.; Awasthi, S.K.; Wang, Q.; Ren, X.; Zhao, J.; Zhang, Z. The behavior of antibiotic resistance genes and their associations with bacterial community during poultry manure composting. Bioresour. Technol. 2019, 280, 70–78. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, Y.; Cheng, D.; Xue, J.; Wakelin, S.; Li, Z. Dynamics of bacterial composition and the fate of antibiotic resistance genes and mobile genetic elements during the co-composting with gentamicin fermentation residue and lovastatin fermentation residue. Bioresour. Technol. 2018, 261, 249–256. [Google Scholar] [CrossRef]
- Yue, Y.; Shen, C.; Ge, Y. Biochar accelerates the removal of tetracyclines and their intermediates by altering soil properties. J. Hazard. Mater. 2019, 380, 120821. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, L.; Fang, Y.; Jia, X.; Chen, J. High temperatures can effectively degrade residual tetracyclines in chicken manure through composting. J. Hazard. Mater. 2019, 380, 120862. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hu, Y.; Cheng, J.; Chen, Y. Degradation of oxytetracycline (OTC) and nitrogen conversion characteristics using a novel strain. Chem. Eng. J. 2018, 354, 758–766. [Google Scholar] [CrossRef]
- Leng, Y.; Bao, J.; Chang, G.; Zheng, H.; Li, X.; Du, J.; Snow, D.; Li, X. Biotransformation of tetracycline by a novel bacterial strain Stenotrophomonas maltophilia DT1. J. Hazard. Mater. 2016, 318, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, X.; Shen, L.; Li, J.; Yu, R.; Liu, Y.; Qiu, G.; Zeng, W. Whole Genome Sequencing and Comparative Genomics Analyses of Pandoraea sp. XY-2, a New Species Capable of Biodegrade Tetracycline. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Yusoff, M.Z.M.; Hu, A.; Feng, C.; Maeda, T.; Shirai, Y.; Hassan, M.A.; Yu, C.P. Influence of pretreated activated sludge for electricity generation in microbial fuel cell application. Bioresour. Technol. 2013, 145, 90–96. [Google Scholar] [CrossRef]
- Pu, C.; Liu, L.; Yao, M.; Liu, H.; Sun, Y. Responses and successions of sulfonamides, tetracyclines and fluoroquinolones resistance genes and bacterial community during the short-term storage of biogas residue and organic manure under the incubator and natural conditions. Environ. Pollut. 2018, 242, 749–759. [Google Scholar] [CrossRef]
- R Studio Team. RStudio: Integrated Development for R; RStudio Inc.: Boston, MA, USA, 2015; Volume 42, p. 14. [Google Scholar]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Ren, J.; Gong, X. Impact of direct application of biogas slurry and residue in fields: In situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 2017, 344, 441–449. [Google Scholar] [CrossRef]
- Zheng, H.-S.; Guo, W.-Q.; Wu, Q.-L.; Ren, N.-Q.; Chang, J.-S. Electro-peroxone pretreatment for enhanced simulated hospital wastewater treatment and antibiotic resistance genes reduction. Environ. Int. 2018, 115, 70–78. [Google Scholar] [CrossRef]
- Thakur, N.; Kumar, V.; Thakur, S.; Sharma, N.; Sheetal; Bhalla, T.C. Biotransformation of 4-hydroxyphenylacetonitrile to 4-hydroxyphenylacetic acid using whole cell arylacetonitrilase of Alcaligenes faecalis MTCC 12629. Process Biochem. 2018, 73, 117–123. [Google Scholar] [CrossRef]
- Jiang, Y.; Wen, J.; Bai, J.; Jia, X.; Hu, Z. Biodegradation of phenol at high initial concentration by Alcaligenes faecalis. J. Hazard. Mater. 2007, 147, 672–676. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, C.; Xu, L.; Zhao, C.; Liang, H.; Qiu, L. Biodegradation of nicosulfuron by a novel Alcaligenes faecalis strain ZWS11. J. Environ. Sci. 2015, 35, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Yao, H.; Wang, H.; Ren, J.; Yu, X. Comparison of ozone and thermal hydrolysis combined with anaerobic digestion for municipal and pharmaceutical waste sludge with tetracycline resistance genes. Water Res. 2016, 99, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, R.; Dai, Q.; Jin, D.; Wang, Y.; Chao, W. Complete degradation of di-n-octyl phthalate by biochemical cooperation between Gordonia sp. strain JDC-2 and Arthrobacter sp. strain JDC-32 isolated from activated sludge. J. Hazard. Mater. 2009, 176, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Jiao, Y.; Chen, X.; Zhou, L.; Guo, K.; Ge, F.; Wu, J. Biodegradation of 4-chloronitrobenzene by biochemical cooperation between Sphingomonas sp. strain CNB3 and Burkholderia sp. strain CAN6 isolated from activated sludge. Chemosphere 2013, 91, 1243–1249. [Google Scholar] [CrossRef]
- Li, Z.; Schulz, L.; Ackley, C.; Fenske, N. Adsorption of tetracycline on kaolinite with pH-dependent surface charges. J. Colloid Interface Sci. 2018, 351, 254–260. [Google Scholar] [CrossRef]
- Loftin, K.A.; Adams, C.D.; Meyer, M.T.; Surampalli, R. Effects of Ionic Strength, Temperature, and pH on Degradation of Selected Antibiotics. J. Environ. Qual. 2008, 37, 378. [Google Scholar] [CrossRef]
- Peng, F.-J.; Zhou, L.-J.; Ying, G.-G.; Liu, Y.-S.; Zhao, J.-L. Antibacterial activity of the soil-bound antimicrobials oxytetracycline and ofloxacin. Environ. Toxicol. Chem. 2014, 33, 776–783. [Google Scholar] [CrossRef]
- Chessa, L.; Pusino, A.; Garau, G.; Mangia, N.P.; Pinna, M.V. Soil microbial response to tetracycline in two different soils amended with cow manure. Environ. Sci. Pollut. Res. 2015, 23, 5807–5817. [Google Scholar] [CrossRef]
- Avisar, D.; Primor, O.; Gozlan, I.; Mamane, H. Sorption of Sulfonamides and Tetracyclines to Montmorillonite Clay. Water Air Soil Pollut. 2009, 209, 439–450. [Google Scholar] [CrossRef]
- Yan, W.; Guo, Y.; Xiao, Y.; Wang, S.; Ding, R.; Jiang, J.; Gang, H.; Wang, H.; Yang, J.; Zhao, F. The changes of bacterial communities and antibiotic resistance genes in microbial fuel cells during long-term oxytetracycline processing. Water Res. 2018, 142, 105–114. [Google Scholar] [CrossRef]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wang, Y.; Zhou, B.; Lin, X. Long-term application of fresh and composted manure increase tetracycline resistance in the arable soil of eastern China. Sci. Total Environ. 2015, 506–507, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhang, T. Occurrence, Abundance, and Diversity of Tetracycline Resistance Genes in 15 Sewage Treatment Plants across China and Other Global Locations. Environ. Sci. Technol. 2018, 45, 2598–2604. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Lu, X.; Zhang, J.; Sui, Q.; Wang, R.; Chen, M.; Wei, Y. Occurrence of antibiotic resistance genes and mobile genetic elements in enterococci and genomic DNA during anaerobic digestion of pharmaceutical waste sludge with different pretreatments. Bioresour. Technol. 2017, 235, 316–324. [Google Scholar] [CrossRef] [PubMed]


| Groups | TRGs | MGEs | |||
|---|---|---|---|---|---|
| Cellular Protection | Efflux Pump | Unknown | Transposase | Integrase | |
| B30-O | 100% | 94.87% | 83.33% | 95.83% | 100% |
| B30-T | 100% | 92.31% | 100 % | 91.67% | 100% |
| Y30-O | 90.90% | 84.61% | 100% | 100% | 50% |
| Y30-T | 81.82% | 76.92% | 100 % | 100% | 50% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Gu, Y.; Wu, X.; Zhou, X.; Zhou, H.; Amanze, C.; Shen, L.; Zeng, W. Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation. Microorganisms 2020, 8, 292. https://doi.org/10.3390/microorganisms8020292
Wu X, Gu Y, Wu X, Zhou X, Zhou H, Amanze C, Shen L, Zeng W. Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation. Microorganisms. 2020; 8(2):292. https://doi.org/10.3390/microorganisms8020292
Chicago/Turabian StyleWu, Xueling, Yichao Gu, Xiaoyan Wu, Xiangyu Zhou, Han Zhou, Charles Amanze, Li Shen, and Weimin Zeng. 2020. "Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation" Microorganisms 8, no. 2: 292. https://doi.org/10.3390/microorganisms8020292
APA StyleWu, X., Gu, Y., Wu, X., Zhou, X., Zhou, H., Amanze, C., Shen, L., & Zeng, W. (2020). Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation. Microorganisms, 8(2), 292. https://doi.org/10.3390/microorganisms8020292




