Comparative Analysis of Fecal Microbiota in Grasscutter (Thryonomys swinderianus) and Other Herbivorous Livestock in Ghana
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Analysis of Fecal Microbiota by 16S rRNA Metagenomics Sequencing
2.3. Statistical Analysis
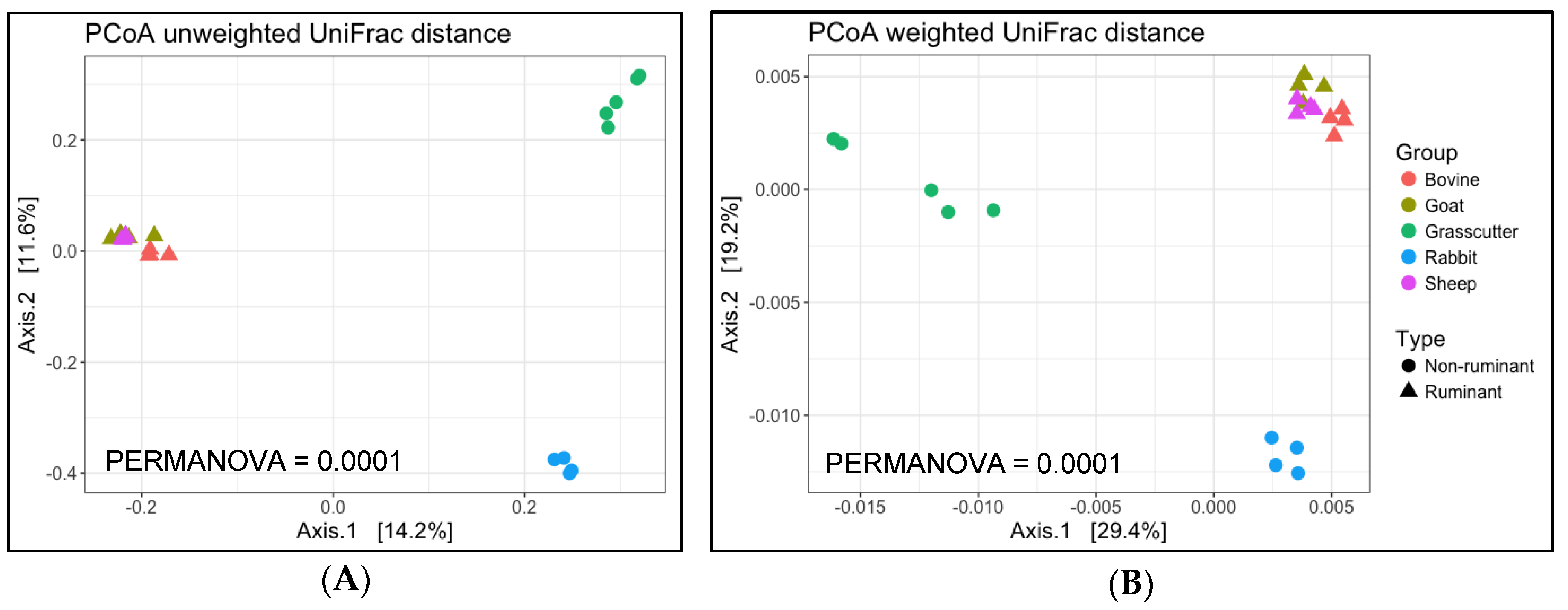
3. Results
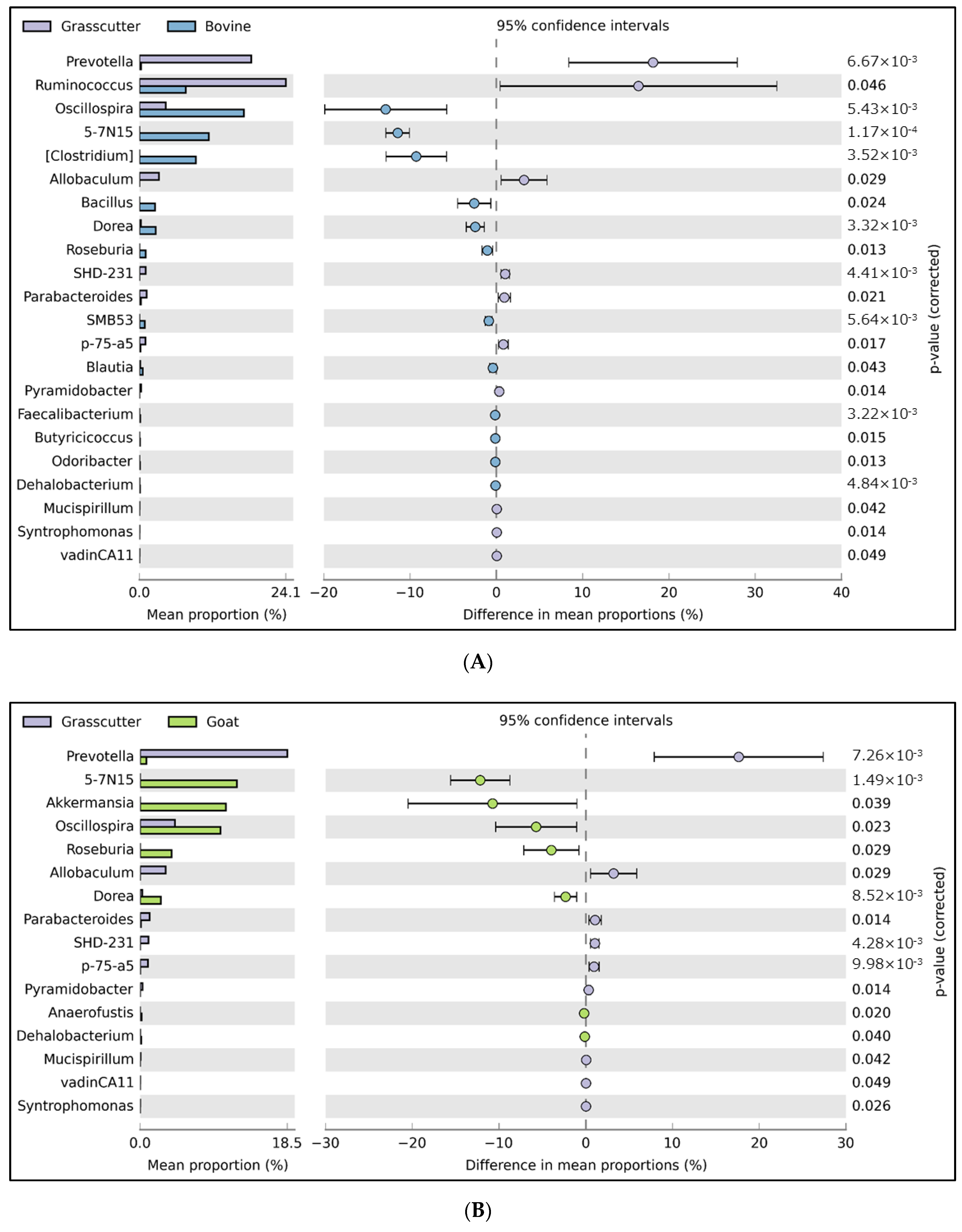
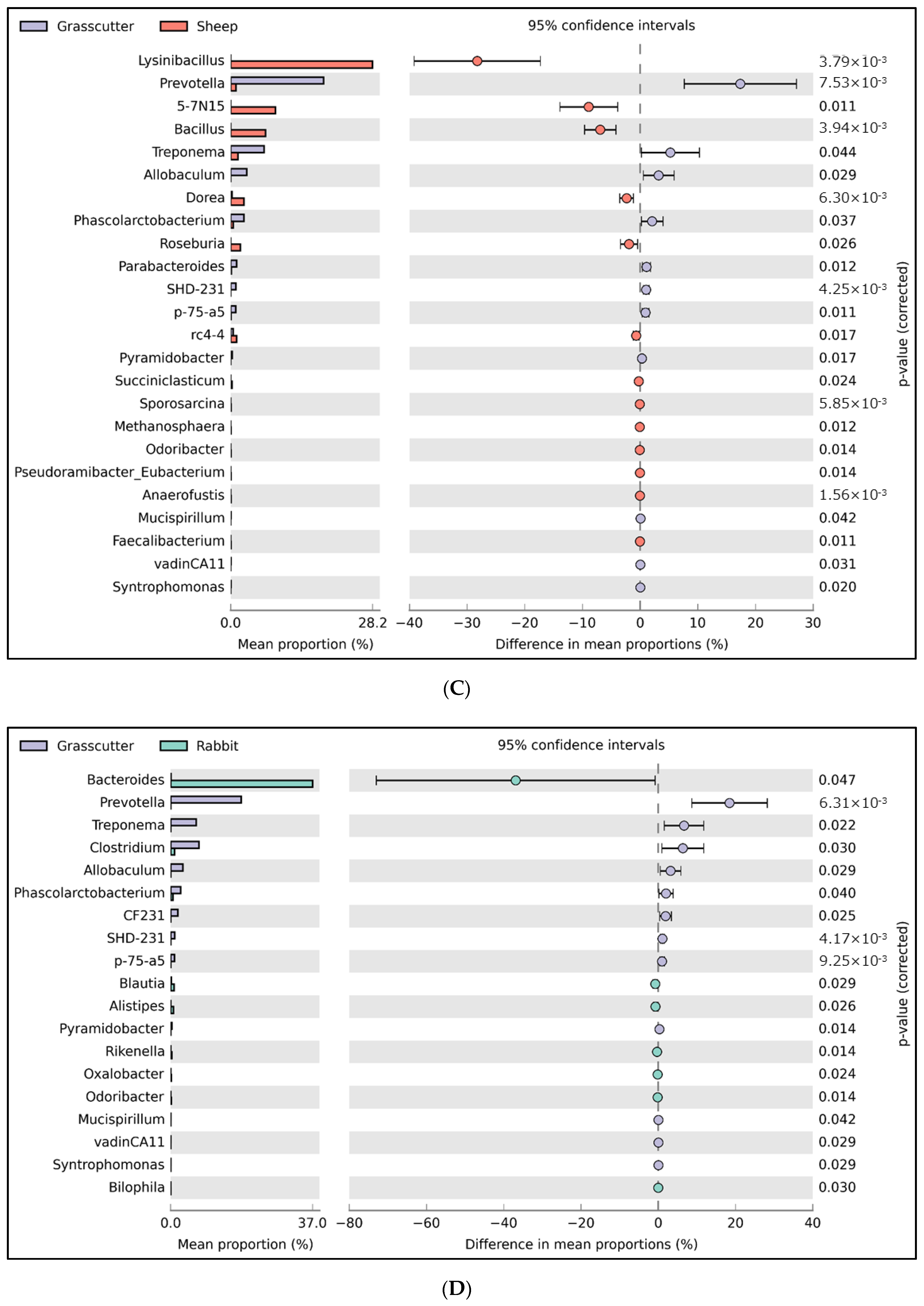
3.1. Relative Abundance of Fecal Microbiota
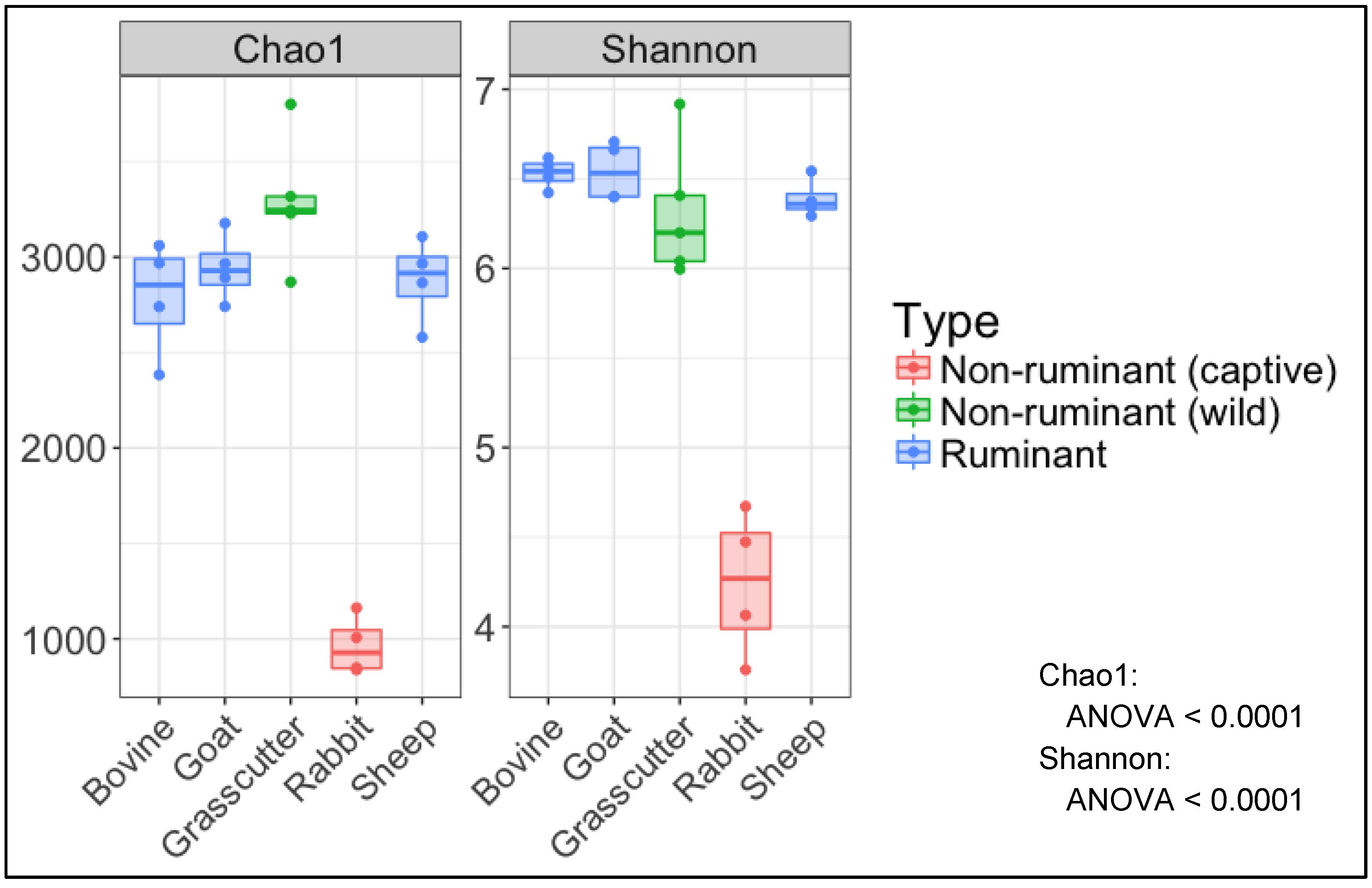
3.2. Analysis of Microbial Diversity for between Animals
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects Volume I: Comprehensive Tables; United Nations: New York, NY, USA, 2019; pp. 22–33. [Google Scholar]
- Saaka, M.; Larbi, A.; Hoeschle-Zeledon, I.; Appiah, B. Child Malnutrition in Northern Ghana: Evidence, Factors and Recommendations from a New Study; International Institute of Tropical Agriculture: Ibadan, Nigeria, 2015; pp. 1–22. [Google Scholar]
- Ohene-Adjei, S.; Bediako, N.A. What is meat in Ghana? Anim. Front. 2017, 7, 60–62. [Google Scholar] [CrossRef]
- McNamara, J.; Fa, J.E.; Ntiamoa-Baidu, Y. Understanding drivers of urban bushmeat demand in a Ghanaian market. Biol. Conserv. 2019, 239, 108291. [Google Scholar] [CrossRef]
- Nadathur, S.R.; Wanasundara, J.P.D.; Scanlin, L. Sustainable Protein Sources; Elsevier: London, UK, 2017; pp. 1–19. [Google Scholar]
- Jori, F.; Mensah, G.A.; Adjanohoun, E. Grasscutter production: An example of rational exploitation of wildlife. Biodivers. Conserv. 1995, 4, 257–265. [Google Scholar] [CrossRef]
- Adenyo, C.; Ogden, R.; Kayang, B.; Onuma, M.; Nakajima, N.; Inoue-Murayama, M. Genome-wide DNA markers to support genetic management for domestication and commercial production in a large rodent, the Ghanaian grasscutter (Thryonomys swinderianus). Anim. Genet. 2017, 48, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Kobayashi, A.; Tsuchida, S.; Yamada, T.; Murata, K.; Nakamura, H.; Ushida, K. Cecal microbiome analyses on wild Japanese rock ptarmigans (Lagopus muta japonica) reveals high level of coexistence of lactic acid bacteria and lactate-utilizing bacteria. Microorganisms 2018, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Kakooza, S.; Mbehang Nguema, P.P.; Wampande, E.M.; Ushida, K. Characteristics of gorilla-specific Lactobacillus isolated from captive and wild gorillas. Microorganisms 2018, 6, 86. [Google Scholar] [CrossRef]
- Pluske, J.R.; Turpin, D.L.; Kim, J.C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2018, 4, 187–196. [Google Scholar] [CrossRef]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Matsuura, Y.; Watanabe, H.; Fujishima, M.; Brune, A. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. USA 2018, 115, E11996–E12004. [Google Scholar] [CrossRef]
- Sakaguchi, E.; Itoh, H.; Uchida, S.; Horigome, T. Comparison of fibre digestion and digesta retention time between rabbits, guinea-pigs, rats and hamsters. Br. J. Nutr. 1987, 58, 149–158. [Google Scholar] [CrossRef]
- Stevens, C.E.; Hume, I.D. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol. Rev. 1998, 78, 393–428. [Google Scholar] [CrossRef]
- Takahashi, T.; Sakaguchi, E. Transport of bacteria across and along the large intestinal lumen of guinea pigs. J. Comp. Physiol. B 2006, 176, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Min, X.; Nishiyama, A.; Sakaguchi, E. Effect of fructo-oligosaccharide on nitrogen utilization in guinea pigs. Anim. Sci. J. 2013, 84, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- De Jesús-Laboy, K.M.; Godoy-Vitorino, F.; Piceno, Y.M.; Tom, L.M.; Pantoja-Feliciano, I.G.; Rivera-Rivera, M.J.; Andersen, G.L.; Domínguez-Bello, M.G. Comparison of the fecal microbiota in feral and domestic goats. Genes 2012, 3, 1–18. [Google Scholar] [CrossRef]
- Rudi, K.; Moen, B.; Sekelja, M.; Frisli, T.; Lee, M.R.F. An eight-year investigation of bovine livestock fecal microbiota. Vet. Microbiol. 2012, 160, 369–377. [Google Scholar] [CrossRef]
- Chen, S.Y.; Deng, F.; Jia, X.; Liu, H.; Zhang, G.W.; Lai, S.J. Gut microbiota profiling with differential tolerance against the reduced dietary fibre level in rabbit. Sci. Rep. 2019, 9, 288. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, H.; Wei, Q.; Zhao, C.; Yang, X.; Wu, X.; Xia, T.; Liu, G.; Zhang, L.; Gao, Y.; et al. Comparative analyses of fecal microbiota in European mouflon (Ovis orientalis musimon) and blue sheep (Pseudois nayaur) living at low or high altitudes. Front. Microbiol. 2019, 10, 1735. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Tokuda, G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 2013, 31, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Abecia, L.; Fondevila, M.; Balcells, J.; McEwan, N.R. The effect of lactating rabbit does on the development of the caecal microbial community in the pups they nurture. J. Appl. Microbiol. 2007, 103, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Arrazuria, R.; Pérez, V.; Molina, E.; Juste, R.A.; Khafipour, E.; Elguezabal, N. Diet induced changes in the microbiota and cell composition of rabbit gut associated lymphoid tissue (GALT). Sci. Rep. 2018, 8, 14103. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Galilea, M.; Piles, M.; Viñas, M.; Rafel, O.; González-Rodríguez, O.; Guivernau, M.; Sánchez, J.P. Rabbit microbiota changes throughout the intestinal tract. Front. Microbiol. 2018, 9, 2144. [Google Scholar] [CrossRef]
- Sakaguchi, E.; Kaizu, K.; Nakamichi, M. Fibre digestion and digesta retention from different physical forms of the feed in the rabbit. Comp. Biochem. Physiol. A 1992, 102, 559–563. [Google Scholar] [CrossRef]
- Sakaguchi, E. Digestive strategies of small hindgut fermenters. Anim. Sci. J. 2003, 74, 327–337. [Google Scholar] [CrossRef]
- Xiao, L.; Xiao, M.; Jin, X.; Kawasaki, K.; Ohta, N.; Sakaguchi, E. Transfer of blood urea nitrogen to cecal microbial nitrogen is increased by mannitol feeding in growing rabbits fed timothy hay diet. Animal 2012, 6, 1757–1763. [Google Scholar] [CrossRef]
- Kawasaki, K.; Min, X.; Li, X.; Hasegawa, E.; Sakaguchi, E. Transfer of blood urea nitrogen to cecal microbial nitrogen is increased by fructo-oligosaccharide feeding in guinea pigs. Anim. Sci. J. 2015, 86, 77–82. [Google Scholar] [CrossRef]
- Crowley, E.J.; King, J.M.; Wilkinson, T.; Worgan, H.J.; Huson, K.M.; Rose, M.T.; McEwan, N.R. Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS ONE 2017, 12, e0165779. [Google Scholar] [CrossRef]
- Li, H.; Qu, J.; Li, T.; Yao, M.; Li, J.; Li, X. Gut microbiota may predict host divergence time during Glires evolution. FEMS Microbiol. Ecol. 2017, 93, fix009. [Google Scholar] [CrossRef] [PubMed]
- O’ Donnell, M.M.; Harris, H.M.B.; Ross, R.P.; O’Toole, P.W. Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals. MicrobiologyOpen 2017, 6, e00509. [Google Scholar] [CrossRef] [PubMed]
- Wogar, G.S.I. Performance of gestating grasscutters (Thryonomys swinderianus) fed cassava-based diets with graded protein levels. Asian J. Anim. Sci. 2011, 5, 373–380. [Google Scholar] [CrossRef]
- Marani, S.A.M. Effects of pelleted feed on the performance of the grasscutter (Thryonomys swinderianus). J. Agric. Sci. Food Res. 2018, 9, 218. [Google Scholar]
- Avenant, N.L.; Power, J.; MacFadyen, D.; Child, M.F. A conservation assessment of Thryonomys swinderianus. In The Red List of Mammals of South Africa, Swaziland and Lesotho; Child, M.F., Roxburgh, L., Do Linh San, E., Raimondo, D., Davies-Mostert, H.T., Eds.; South African National Biodiversity Institute and Endangered Wildlife Trust: Midrand, South Africa, 2016; pp. 1–6. [Google Scholar]
- Mulungu, L.S. Control of rodent pests in maize cultivation: The case of Africa. In Achieving Sustainable Cultivation of Maize; Watson, D., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2017; Volume 2, pp. 317–338. [Google Scholar]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhou, H.; Li, F.; Zhang, N.; Zhu, Y. Effect of dietary fiber levels on bacterial composition with age in the cecum of meat rabbits. MicrobiologyOpen 2019, 8, e00708. [Google Scholar] [CrossRef] [PubMed]
| Non-ruminant | Ruminant | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grasscutter | Rabbit | Cattle | Goat | Sheep | ||||||
| No. | Taxonomy 1 | (%) | Taxonomy | (%) | Taxonomy | (%) | Taxonomy | (%) | Taxonomy | (%) |
| 1 | Bacteroidales | 11.59 | Bacteroides | 20.89 | Ruminococcaceae | 28.27 | Ruminococcaceae | 31.29 | Ruminococcaceae | 25.14 |
| 2 | Ruminococcaceae | 10.44 | Clostridiales | 16.62 | Clostridiales | 9.79 | Clostridiales | 11.12 | Clostridiales | 9.85 |
| 3 | Clostridiales | 9.79 | Lachnospiraceae | 10.41 | Bacteroidales | 7.64 | Bacteroidales | 7.75 | Lysinibacillus | 8.74 |
| 4 | Ruminococcus | 9.15 | Ruminococcus | 7.71 | Oscillospira | 4.69 | Lachnospiraceae | 4.97 | Bacteroidales | 7.21 |
| 5 | Lachnospiraceae | 6.71 | Anaeroplasma | 6.29 | Lachnospiraceae | 4.10 | Bacillales | 4.16 | Lachnospiraceae | 4.19 |
| 6 | Prevotella | 6.66 | Akkermansia | 5.32 | 5-7N15 | 3.17 | Christensenellaceae | 3.64 | Rikenellaceae | 3.41 |
| 7 | Fibrobacter | 3.56 | Oscillospira | 4.73 | Clostridium | 2.73 | Lysinibacillus | 3.50 | Bacillales | 3.20 |
| 8 | RF16 | 3.24 | Rikenellaceae | 3.84 | [Clostridium] | 2.55 | 5-7N15 | 3.50 | Ruminococcus | 3.07 |
| 9 | Treponema | 2.35 | Bacillus | 3.00 | Rikenellaceae | 2.43 | Akkermansia | 3.01 | 5-7N15 | 2.75 |
| 10 | S24-7 | 1.99 | Ruminococcaceae | 2.99 | [Mogibacteriaceae] | 2.40 | Oscillospira | 2.92 | Christensenellaceae | 2.49 |
| Total | 65.48 | 81.80 | 67.77 | 75.87 | 70.03 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawasaki, K.; Ohya, K.; Omatsu, T.; Katayama, Y.; Takashima, Y.; Kinoshita, T.; Odoi, J.O.; Sawai, K.; Fukushi, H.; Ogawa, H.; et al. Comparative Analysis of Fecal Microbiota in Grasscutter (Thryonomys swinderianus) and Other Herbivorous Livestock in Ghana. Microorganisms 2020, 8, 265. https://doi.org/10.3390/microorganisms8020265
Kawasaki K, Ohya K, Omatsu T, Katayama Y, Takashima Y, Kinoshita T, Odoi JO, Sawai K, Fukushi H, Ogawa H, et al. Comparative Analysis of Fecal Microbiota in Grasscutter (Thryonomys swinderianus) and Other Herbivorous Livestock in Ghana. Microorganisms. 2020; 8(2):265. https://doi.org/10.3390/microorganisms8020265
Chicago/Turabian StyleKawasaki, Kiyonori, Kenji Ohya, Tsutomu Omatsu, Yukie Katayama, Yasuhiro Takashima, Tsuyoshi Kinoshita, Justice Opare Odoi, Kotaro Sawai, Hideto Fukushi, Hirohito Ogawa, and et al. 2020. "Comparative Analysis of Fecal Microbiota in Grasscutter (Thryonomys swinderianus) and Other Herbivorous Livestock in Ghana" Microorganisms 8, no. 2: 265. https://doi.org/10.3390/microorganisms8020265
APA StyleKawasaki, K., Ohya, K., Omatsu, T., Katayama, Y., Takashima, Y., Kinoshita, T., Odoi, J. O., Sawai, K., Fukushi, H., Ogawa, H., Inoue-Murayama, M., Mizutani, T., Adenyo, C., Matsumoto, Y., & Kayang, B. (2020). Comparative Analysis of Fecal Microbiota in Grasscutter (Thryonomys swinderianus) and Other Herbivorous Livestock in Ghana. Microorganisms, 8(2), 265. https://doi.org/10.3390/microorganisms8020265





