Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strain
2.2. Propolis
2.3. Chemicals and Solvents
2.4. Extraction of Phenolic Compounds from Crude Propolis
2.5. Spectrophotometric Analysis of Total Phenolics
2.6. Sample Preparation for HPLC Analysis
2.7. HPLC Analysis of Phenolics in Propolis Extracts
2.8. Minimal Inhibitory Concentration (MIC) Assay
2.9. Assessment of Propolis Effects on Microbial Growth and Early Biofilm Formation
2.10. Assessment of Phenazines and Propolis Polyphenols in Cell-Free Supernatants
2.11. Assessment of Propolis Effects on eDNA Release
2.12. Statistical Analysis
3. Results
3.1. Total Phenolic Compounds in Propolis Extracts
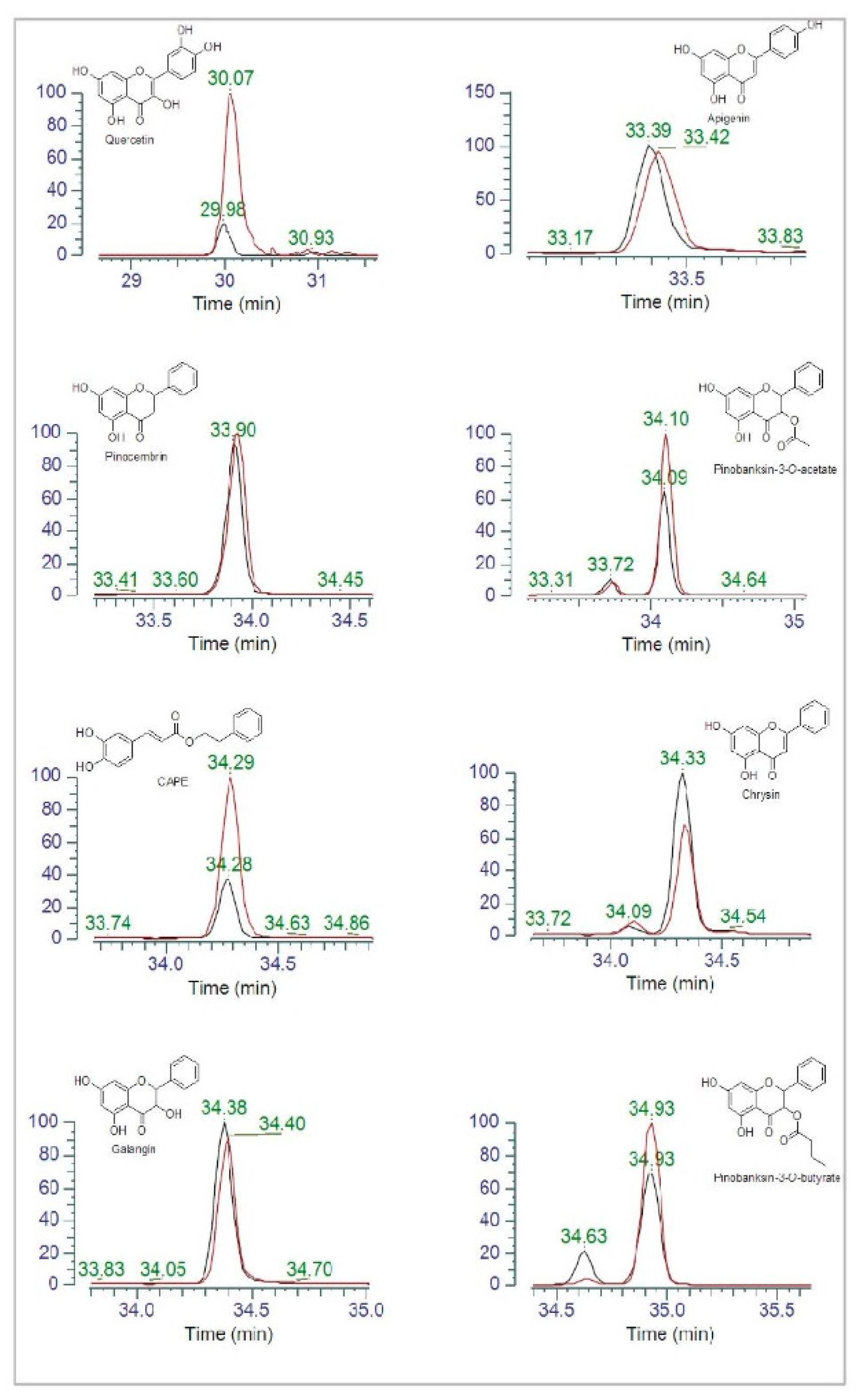
3.2. HPLC Analysis of Polyphenols in Propolis Extracts
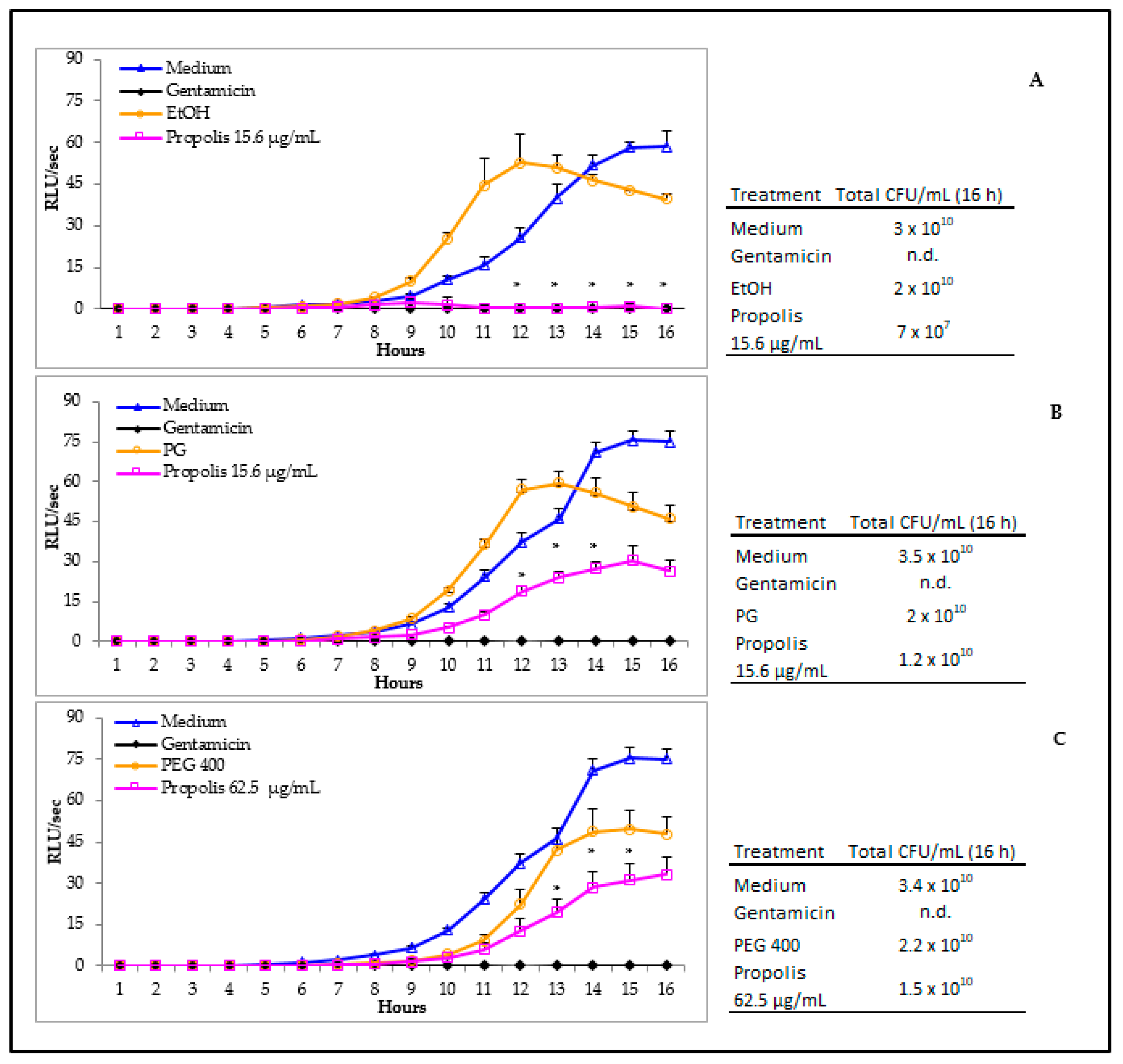
3.3. Antibacterial Activity of Propolis
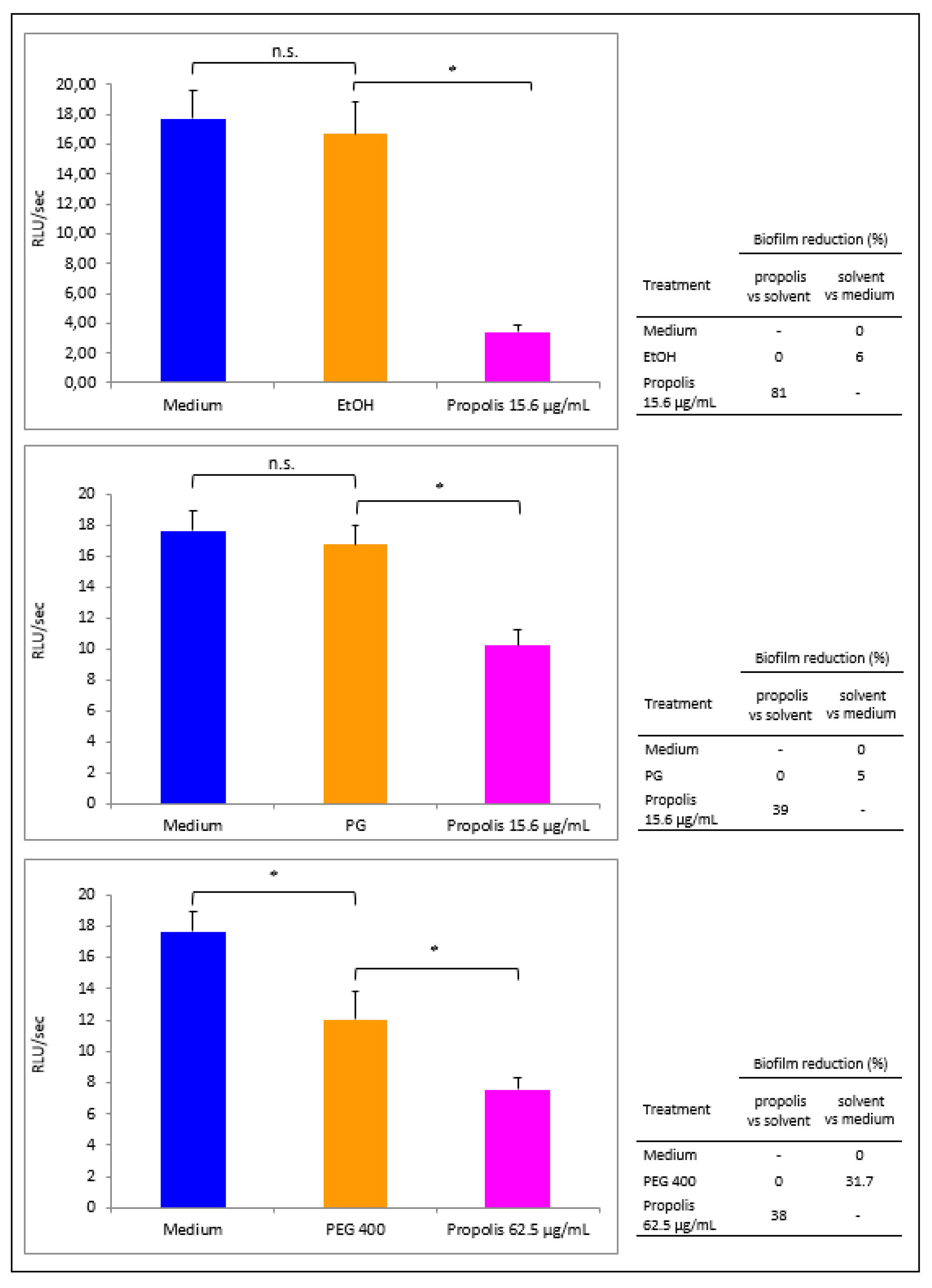
3.4. Propolis Effects on P. aeruginosa Early Biofilm Formation
3.5. Propolis Effects on Phenazines Release by P. aeruginosa
3.6. Propolis Effects on eDNA release by P. aeruginosa
3.7. Polyphenol Content in Propolis Exposed or Not to P. aeruginosa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tsuneda, S.; Aikawa, H.; Hayashi, H.; Yuasa, A.; Hirata, A. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol. Lett. 2003, 223, 287–292. [Google Scholar] [CrossRef]
- Das, T.; Sharma, P.K.; Busscher, H.J.; Van der Mei, H.C.; Krom, B.P. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Mazenod, L.C.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Smith, W.D.; Bardin, E.; Cameron, L.; Edmondson, C.L.; Farrant, K.V.; Martin, I.; Murphy, R.A.; Soren, O.; Turnbull, A.R.; Wierre-Gore, N.; et al. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol. Lett. 2017, 364, fnx121. [Google Scholar] [CrossRef]
- Nord, C.E.; Sjöberg, L.; Wadström, T. Pseudomonas aeruginosa in oral infections. Acta Odontol. Scand. 1972, 30, 371–381. [Google Scholar] [CrossRef]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Bonsall, R.F.; Delaney, S.M.; Soule, M.J.; Phillips, G.; Thomashow, L.S. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6454–6465. [Google Scholar] [CrossRef]
- Das, T.; Mike, M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e46718. [Google Scholar]
- Pericolini, E.; Colombari, B.; Ferretti, G.; Iseppi, R.; Ardizzoni, A.; Girardis, M.; Sala, A.; Peppoloni, S.; Blasi, E. Real-time monitoring of Pseudomonas aeruginosa biofilm formation on endotracheal tubes in vitro. BMC Microbiol. 2018, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, S.; Capasso, F. Propolis, an old remedy used in modern medicine. Fitoterapia 2002, 73, S1–S6. [Google Scholar] [CrossRef]
- Gambichler, T.; Boms, S.; Freitag, M. Contact dermatitis and other skin conditions in instrumental musicians. BMC Dermatol. 2004, 4, 3. [Google Scholar] [CrossRef]
- Toreti, V.C.; Sato, H.H.; Pastore, G.M.; Park, Y.K. Recent progress of propolis for its biological and chemical compositions and its botanical origin. Evid. Based Complement. Altern. Med. 2013, 2013, 1–13. [Google Scholar] [CrossRef]
- Bankova, V. Chemical diversity of propolis and the problem of standardization. J. Ethnopharmacol. 2005, 100, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Dantas Silva, R.P.; Machado, B.A.; Barreto, G.A.; Costa, S.S.; Andrade, L.N.; Amaral, R.G.; Carvalho, A.A.; Padilha, F.F.; Barbosa, J.D.; Umsza-Guez, M.A. Antioxidant, antimicrobial, antiparasitic, and cytotoxic properties of various Brazilian propolis extracts. PLoS ONE 2017, 12, e0172585. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Naseem, M.; Zafar, M.S.; Najeeb, S.; Zohaib, S. Propolis: A natural biomaterial for dental and oral healthcare. J. Dent. Res. Dent. Clin. Dent. Prospect. 2017, 11, 265–274. [Google Scholar]
- Ikeno, K.; Ikeno, T.; Miyazawa, C. Effects of propolis on dental caries in rats. Caries Res. 1991, 25, 347–351. [Google Scholar] [CrossRef]
- Özan, F.; Polat, Z.A.; Er, K.; Özan, U.; Değer, O. Effect of propolis on survival of periodontal ligament cells: New storage media for avulsed teeth. J. Endod. 2007, 33, 570–573. [Google Scholar] [CrossRef]
- Abdulrhman, M.; Samir, E.N.; Ahmed, A.D.; Saeid, E.R. Honey and a mixture of honey, beeswax, and olive oil–propolis extract in treatment of chemotherapy-induced oral mucositis: A randomized controlled pilot study. Pediatr. Hematol. Oncol. 2012, 29, 285–292. [Google Scholar] [CrossRef]
- Murray, M.C.; Worthington, H.V.; Blinkhorn, A.S. A study to investigate the effect of a propolis-containing mouthrinse on the inhibition of de novo plaque formation. J. Clin. Periodontol. 1997, 24, 796–798. [Google Scholar] [CrossRef]
- Meto, A.; Meto, A.; Bimbari, B.; Shytaj, K.; Mutlu, Ö. Antiinflammatory and regenerative effects of Albanian propolis in experimental vital amputations. Eur. J. Prosthodont. Restor. Dent. 2016, 24, 145–151. [Google Scholar]
- Sabir, A. The healing actions of propolis on direct pulp capping treatment: A review. J. Dentomaxillofacial Sci. 2016, 1, 188. [Google Scholar] [CrossRef][Green Version]
- Kayaoglu, G.; Ömürlü, H.; Akca, G.; Gürel, M.; Gençay, Ö.; Sorkun, K.; Salih, B. Antibacterial activity of Propolis vs. conventional endodontic disinfectants against Enterococcus faecalis in infected dentinal tubules. J. Endod. 2011, 37, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Król, W.; Czuba, Z.; Szliszka, E.; Mertas, A.; Skaba, D. Biological and therapeutic properties of ethanol Brazilian green propolis extract. Inz. Biomateriałow Stomatol. 2010, 7, 2–4. [Google Scholar]
- Mattigatti, S.; Ratnakar, P.; Moturi, S.; Varma, S.; Rairam, S. Antimicrobial effect of conventional root canal medicaments vs. propolis against Enterococcus faecalis, Staphylococcus aureus and Candida albicans. J. Contemp. Dent. Pract. 2012, 13, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.; Jim, W.; Su, L.; Chung, C.; Lin, C.; Huo, C.; Tseng, J.; Huang, S.; Lai, C.; Chen, B. Caffeic acid phenethyl ester is a potential therapeutic agent for oral cancer. Int. J. Mol. Sci. 2015, 16, 10748–10766. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, C.C.; Fernandes, L.M.; Silva, M.L.; Luz, D.A.; Gomes, A.R.; Monteiro, M.C.; Machado, C.S.; Torres, Y.R.; de Lira, T.O.; Ferreira, A.G.; et al. Neurobehavioral and antioxidant effects of Ethanolic extract of yellow Propolis. Oxidative Med. Cell Longev. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rosseto, H.C.; Toledo, L.A.S.; Francisco, L.M.B.; Esposito, E.; Lim, Y.; Valacchi, G.; Cortesi, R.; Bruschi, M.L. Nanostructured lipid systems modified with waste material of propolis for wound healing: Design, in vitro and in vivo evaluation. Colloids Surf. B Biointerfaces 2017, 158, 441–452. [Google Scholar] [CrossRef]
- Choi, K.H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas Aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A., Jr. Colorunetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2011. [Google Scholar]
- Pellati, F.; Prencipe, F.P.; Bertelli, D.; Benvenuti, S. An efficient chemical analysis of phenolic acids and flavonoids in raw propolis by microwave-assisted extraction combined with high-performance liquid chromatography using the fused-core technology. J. Pharm. Biomed. Anal. 2013, 81–82, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Sforcin, J.M. Biological properties and therapeutic applications of propolis. Phytother. Res. 2016, 30, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Kędzia, B.; Hołderna-Kędzia, E. The antibiotic activity of native and European propolis. Post. Fitoter. 2013, 2, 97–107. [Google Scholar]
- Preetha, R.; Jose, S.; Prathapan, S.; Vijayan, K.K.; Jayaprakash, N.S.; Philip, R.; Bright-Singh, I.S. An inhibitory compound produced by Pseudomonas with effectiveness on Vibriyo harveyi. Aquac. Res. 2010, 41, 1452–1461. [Google Scholar]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; Hoiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1- hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Ramos, I.; Dietrich, L.E.P.; Pricewhelan, A.; Newman, D.K. Phenazine affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 2010, 161, 187–191. [Google Scholar] [CrossRef]
- Pricewhelan, A.; Dietrich, L.E.P.; Newman, D.K. Rethinking secondary metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef]
- Orsolic, N.; Knezevic, A.H.; Sver, L.; Terzic, S.; Basic, I. Immunomodulatory and antimetastatic action of propolis and related polyphenolic compounds. J. Ethnopharmacol. 2004, 94, 307–315. [Google Scholar] [CrossRef]
- Sforcin, J.M. Propolis and the immune system: A review. J. Ethnopharmacol. 2007, 113, 1–14. [Google Scholar] [CrossRef]
- Sforcin, J.M.; Bankova, V. Propolis: Is there a potential for the development of new drugs? J. Ethnopharmacol. 2011, 133, 253–260. [Google Scholar] [CrossRef]
- Pellati, F.; Orlandini, G.; Pinetti, D.; Benvenuti, S. HPLC-DAD and HPLC-ESI-MS/MS methods for metabolite profiling of propolis extracts. J. Pharm. Biomed. Anal. 2011, 55, 934–948. [Google Scholar] [CrossRef]
- Boesch-Saadatmandi, C.; Loboda, A.; Wagner, A.E.; Stachurska, A.; Jozkowicz, A.; Dulak, J.; Döring, F.; Wolffram, S.; Rimbach, G. Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: Role ofmiR-155. J. Nutr. Biochem. 2011, 22, 293–299. [Google Scholar] [CrossRef]
- Boesch-Saadatmandi, C.; Wagner, A.E.; Wolffram, S.; Rimbach, G. Effect of quercetin on inflammatory gene expression in mice liver in vivo-role of redox factor 1, miRNA-122 and miRNA-125b. Pharmacol. Res. 2012, 65, 523–530. [Google Scholar] [CrossRef]
- Islam, B.; Khan, S.N.; Haque, I.; Alam, M.; Mushfiq, M.; Khan, A.U. Novel anti-adherence activity of mulberry leaves: Inhibition of Streptococcus mutans biofilm by 1-deoxynojirimycin isolated from Morus alba. J. Antimicrob. Chemother. 2008, 6, 751–757. [Google Scholar] [CrossRef]
- Kabała-Dzik, A.; Rzepecka-Stojko, A.; Kubina, R.; Jastrzębska-Stojko, Ż.; Stojko, R.; Wojtyczka, R.D.; Stojko, J. Comparison of two components of propolis: Caffeic Acid (CA) and Caffeic Acid Phenethyl Ester (CAPE) induce apoptosis and cell cycle arrest of breast cancer cells MDA-MB-23. Molecules 2017, 2, 1554. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, T.; Du, J.; Cui, S.; Yang, F.; Jin, Q. Anti-enterovirus 71 effects of chrysin and its phosphate ester. PLoS ONE 2014, 9, e89668. [Google Scholar] [CrossRef]
- Ouyang, J.; Sun, F.; Feng, W.; Xie, Y.; Ren, L.; Chen, Y. Antimicrobial Activity of Galangin and Its Effects on Murein Hydrolases of Vancomycin-Intermediate Staphylococcus aureus (VISA) Strain Mu50. Chemotherapy 2018, 63, 20–28. [Google Scholar] [CrossRef]
- Veloz, J.J.; Alvear, M.; Salazar, L.A. Antimicrobial and Antibiofilm Activity against Streptococcus mutans of Individual and Mixtures of the Main Polyphenolic Compounds Found in Chilean Propolis. BioMed. Res. Int. 2019, 2019, 7. [Google Scholar] [CrossRef]
- Zheng, Y.Z.; Deng, G.; Chen, D.F.; Liang, Q.; Guo, R.; Fu, Z.M. Theoretical studies on the antioxidant activity of pinobanksin and its ester derivatives: Effects of the chain length and solvent. Food Chem. 2018, 240, 323–329. [Google Scholar] [CrossRef]
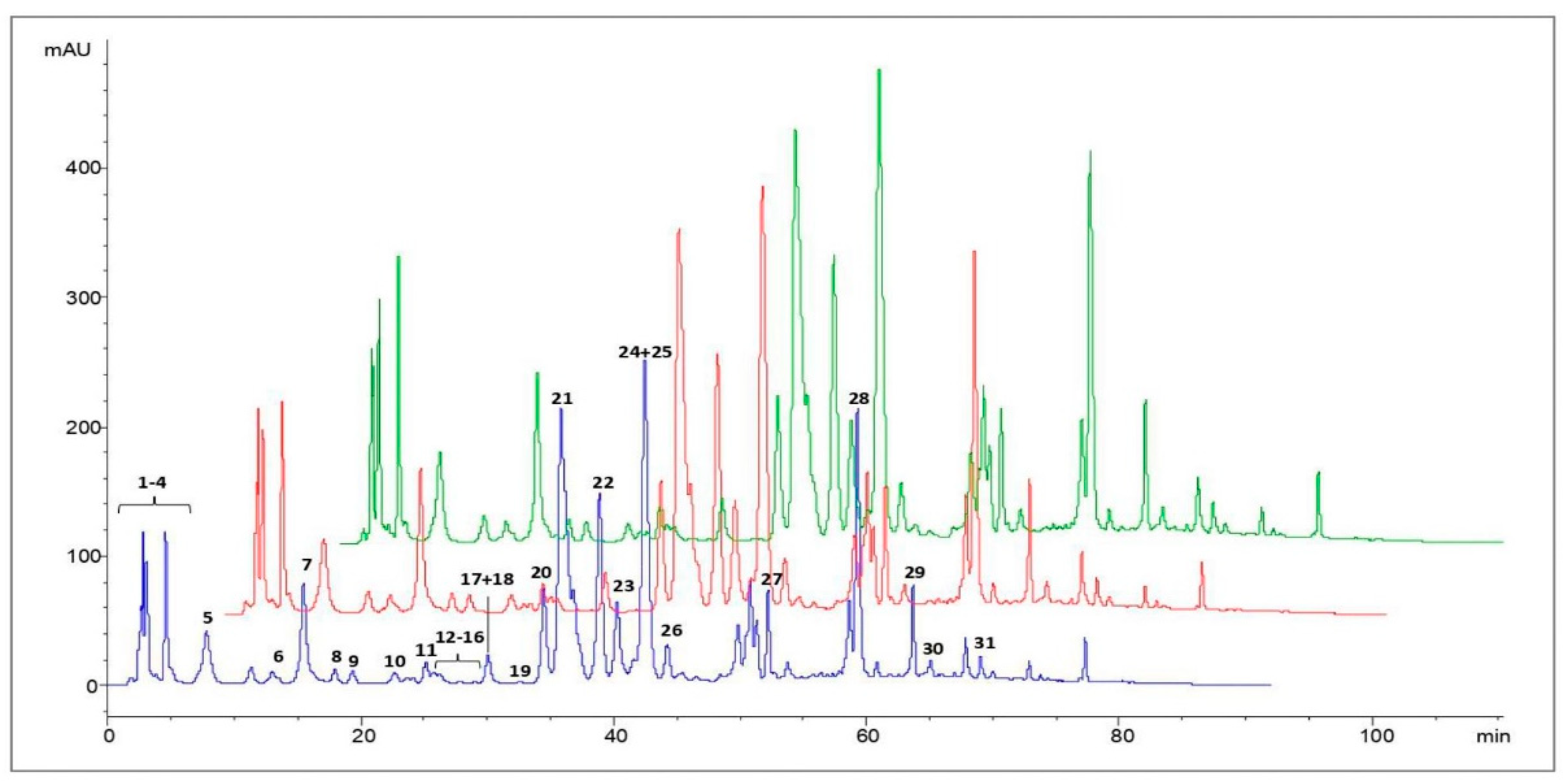
| Peak Number | Compounds | UV λmax(nm) | [M + H]+ | [M − H]− | MS2 Fragments (m/z) |
|---|---|---|---|---|---|
| 1 | Caffeic acid | 298,324 | 179 | 135 | |
| 2 | p-Coumaric acid | 298,310 | 163 | 119 | |
| 3 | Ferulic acid | 298,324 | 193 | 149, 134 | |
| 4 | Isoferulic acid | 296,321 | 193 | 149, 134 | |
| 5 | 3,4-Dimethyl-caffeic acid (DMCA) | 296,322 | 207 | 163, 133 | |
| 6 | Quercetin | 256,372 | 303 | 285, 257, 229, 165, 153, 149 | |
| 7 | Pinobanksin-5-methyl-ether | 288,318 sh | 287 | 269, 241, 152, 91 | |
| 8 | Quercetin-3-methyl-ether | 256,358 | 317 | 302, 165, 153, 137 | |
| 9 | Chrysin-5-methyl-ether | 264,314 | 269 | 254, 167 | |
| 10 | Apigenin | 267,338 | 271 | 253, 153, 119 | |
| 11 | Pinobanksin | 291,330 sh | 273 | 255, 227, 153 | |
| 12 | Isorhamnetin | 255,372 | 317 | 302, 285, 177, 153 | |
| 13 | Luteolin-methyl-ether | 266,350 | 301 | 286, 217 | |
| 14 | Quercetin-dimethyl-ether | 254,356 | 331 | 316, 301, 299 | |
| 15 | Galangin-5-methyl-ether | 260,302 sh, 352 | 285 | 270, 239, 167 | |
| 16 | Pinobanksin-5-methyl-ether-3-O-acetate | 288,326 | 329 | 287, 241 | |
| 17 | Cinnamilidenacetic acid | 312 | 175 | 157, 129 | |
| 18 | Quercetin-7-methyl-ether | 256,372 | 317 | 302, 271, 243, 179, 167 | |
| 19 | Quercetin-dimethyl-ether | 256,357 | 331 | 316, 299 | |
| 20 | Caffeic acid prenyl ester | 298,326 | 247 | 179, 135 | |
| 21 | Chrysin | 268,314 sh | 255 | 209, 153, 129 | |
| 22 | Pinocembrin | 290,330 sh | 257 | 215, 153, 131, 103 | |
| 23 | Galangin | 260,308 sh, 360 | 271 | 165, 153, 105 | |
| 24 | Caffeic acid phenylethyl ester (CAPE) | 298,328 | 283 | 179, 135 | |
| 25 | Pinobanksin-3-O-acetate | 294,332 sh | 315 | 273, 255, 227, 153 | |
| 26 | Methoxy-chrysin | 266,310 sh, 340 sh | 285 | 270, 257, 242 | |
| 27 | Pinobanksin-3-O-propionate | 292,330 sh | 329 | 273, 255, 227, 153 | |
| 28 | Pinobanksin-3-O-butyrate * | 268,310 sh | 343 | 273, 255, 227, 153 | |
| 29 | Pinobanksin-3-O-pentanoate * | 292,332 sh | 357 | 273, 255, 227, 153 | |
| 30 | Pinobanksin-3-O-hexanoate * | 282 | 371 | 273, 255, 227, 153 | |
| 31 | p-Methoxy cinnamic acid cinnamyl ester | 278 | 295 | 149 |
| Phenazines | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| PCA | PYO | 1-OH-PHZ | |||||||
| Reduction (%) | Reduction (%) | Reduction (%) | |||||||
| Treatment | Peak Area | Propolis vs. Medium | Propolis vs. Solvent | Peak Area | Propolis vs. Medium | Propolis vs. Solvent | Peak Area | Propolis vs. Medium | Propolis vs. Solvent |
| Medium | 2.7 × 109 | 0 | - | 5.1 × 109 | 0 | - | 4.1 × 107 | 0 | - |
| EtOH | 3 × 109 | - | 0 | 5.3 × 109 | - | 0 | 4.4 × 107 | - | 0 |
| Propolis 15.6 µg/mL | 9.7 × 108 | 64 | 68 | 2.3 × 109 | 55 | 56.6 | 1.7 × 107 | 58.5 | 61.4 |
| EtOH | 1.7 × 109 | - | 0 | 3 × 109 | - | 0 | 2.4 × 107 | - | 0 |
| Propolis 31.2 µg/mL | 2.2 × 108 | 92 | 87.1 | 5 × 108 | 90.2 | 83.4 | 3.2 × 106 | 92.2 | 86.7 |
| eDNA | |||
|---|---|---|---|
| Treatment | RFU | Reduction (%) | |
| Propolis vs. Medium | Propolis vs. Solvent | ||
| Medium | 0.121 | 0 | - |
| EtOH | 0.126 | - | 0 |
| Propolis 15.6 µg/mL | 0.091 | 24.8 | 27.8 |
| EtOH | 0.081 | - | 0 |
| Propolis 31.2 µg/mL | 0.068 | 43.8 | 16.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meto, A.; Colombari, B.; Meto, A.; Boaretto, G.; Pinetti, D.; Marchetti, L.; Benvenuti, S.; Pellati, F.; Blasi, E. Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols. Microorganisms 2020, 8, 243. https://doi.org/10.3390/microorganisms8020243
Meto A, Colombari B, Meto A, Boaretto G, Pinetti D, Marchetti L, Benvenuti S, Pellati F, Blasi E. Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols. Microorganisms. 2020; 8(2):243. https://doi.org/10.3390/microorganisms8020243
Chicago/Turabian StyleMeto, Aida, Bruna Colombari, Agron Meto, Giorgia Boaretto, Diego Pinetti, Lucia Marchetti, Stefania Benvenuti, Federica Pellati, and Elisabetta Blasi. 2020. "Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols" Microorganisms 8, no. 2: 243. https://doi.org/10.3390/microorganisms8020243
APA StyleMeto, A., Colombari, B., Meto, A., Boaretto, G., Pinetti, D., Marchetti, L., Benvenuti, S., Pellati, F., & Blasi, E. (2020). Propolis Affects Pseudomonas aeruginosa Growth, Biofilm Formation, eDNA Release and Phenazine Production: Potential Involvement of Polyphenols. Microorganisms, 8(2), 243. https://doi.org/10.3390/microorganisms8020243









