Abstract
Upland soils of tundra function as a constant sink for atmospheric CH4 but the identity of methane oxidizers in these soils remains poorly understood. Methane uptake rates of −0.4 to −0.6 mg CH4-C m−2 day−1 were determined by the static chamber method in a mildly acidic upland soil of the lichen-dominated forested tundra, North Siberia, Russia. The maximal CH4 oxidation activity was localized in an organic surface soil layer underlying the lichen cover. Molecular identification of methanotrophic bacteria based on retrieval of the pmoA gene revealed Upland Soil Cluster Alpha (USCα) as the only detectable methanotroph group. Quantification of these pmoA gene fragments by means of specific qPCR assay detected ~107 pmoA gene copies g−1 dry soil. The pmoA diversity was represented by seven closely related phylotypes; the most abundant phylotype displayed 97.5% identity to pmoA of Candidatus Methyloaffinis lahnbergensis. Further analysis of prokaryote diversity in this soil did not reveal 16S rRNA gene fragments from well-studied methanotrophs of the order Methylococcales and the family Methylocystaceae. The largest group of reads (~4% of all bacterial 16S rRNA gene fragments) that could potentially belong to methanotrophs was classified as uncultivated Beijerinckiaceae bacteria. These reads displayed 96–100 and 95–98% sequence similarity to 16S rRNA gene of Candidatus Methyloaffinis lahnbergensis and “Methylocapsa gorgona” MG08, respectively, and were represented by eight species-level operational taxonomic units (OTUs), two of which were highly abundant. These identification results characterize subarctic upland soils, which are exposed to atmospheric methane concentrations only, as a unique habitat colonized mostly by USCα methanotrophs.
1. Introduction
Methane (CH4) is one of the most impactful greenhouse gases, which has contributed about 20% of the additional radiative forcing accumulated in the lower atmosphere since 1750 [1,2,3,4]. Emissions and concentrations of CH4 are continuing to increase; the current atmospheric CH4 mixing ratio is 1.8 ppmv. Major biological net sinks for atmospheric methane are well aerated upland soils [5,6,7]. Atmospheric CH4 uptake in these soils is due to activity of aerobic methanotrophic bacteria, which utilize methane as a source of energy [8,9,10,11]. Aerobic methanotrophs comprise a number of bacterial genera within the Gamma- and Alphaproteobacteria, as well as the Verrucomicrobia [12]. A key enzyme of the methanotrophic metabolism is particulate methane monooxygenase (pMMO), which is present in most currently described methanotroph species. Accordingly, the pmoA gene coding for the active-site polypeptide of pMMO is the most frequently used molecular marker in cultivation-independent detection of aerobic methanotrophs [13].
The apparent affinity for CH4 measured for pure cultures of most currently described methanotroph species is in the range of 1–10 µm, while the respective values determined for upland soils are in the nanomolar range [14]. Microorganisms that are able to oxidize atmospheric CH4, therefore, are often referred to as “high-affinity” methanotrophs. The identity of high-affinity methane oxidizers in terrestrial ecosystems has been the focus of considerable microbiological research. The first evidence that as-yet-uncultivated methanotroph group is involved in atmospheric CH4 oxidation in upland soils was obtained by both labeling phospholipid fatty acids of methanotrophs with 14CH4 and analyzing the pmoA gene library [15]. The pmoA sequences retrieved in this study could not be affiliated to any of the earlier described methanotrophs and displayed only a distant relationship to pmoA gene from Methylocapsa acidiphila, an alphaproteobacterium isolated from acidic peat [16,17]. This previously unknown group of pmoA sequences, called Upland Soil Cluster Alphaproteobacteria (USCα) after Knief et al. [18], has been recovered from many acidic and pH-neutral boreal upland soils [18,19,20,21,22,23]. These bacteria were also identified as the predominant methanotroph group in acidic carbon-poor cryosols at Axel Heiberg Island in the Canadian high Arctic, which were shown to consistently consume atmospheric methane [24].
The exact phylogenetic position of USCα methanotrophs remained unknown for a long time but their tentative affiliation to the alphaproteobacterial family Beijerinckiaceae was suggested by several metagenomic studies [25,26,27]. Reconstruction of a draft genome of USCα methanotroph enabled the first insights into the metabolic potential and environmental adaptation strategies of these methanotrophs [27]. The use of multilocus sequence analysis suggested placement of the metagenomic assembly obtained in the latter study in the new, Methylocapsa-related genus of the family Beijerinckiaceae, Candidatus Methyloaffinis lahnbergensis.
Recent isolation of a pure culture of the first cultivated member of the USCα clade, strain MG08, provided valuable insights into the physiology and metabolism of these methanotrophs [28]. CH4 oxidation experiments and 13C-single cell isotope analyses proved the ability of strain MG08 to grow at atmospheric concentrations of CH4 and to assimilate carbon from both CH4 and CO2. The genome of strain MG08 encodes the serine cycle for assimilation of carbon from CH4 and CO2, and CO2 fixation through the reductive glycine pathway. This strain also fixes N2 and expresses the genes for a high-affinity hydrogenase and carbon monoxide dehydrogenase, suggesting that oxidation of the atmospheric trace gases, CO and H2, provides additional energy sources [28]. Based on the results of comparative genome analysis, strain MG08 was classified as a member of the genus Methylocapsa and was tentatively named “Methylocapsa gorgona” MG08. Although the PmoA sequence of Methylocapsa gorgona MG08 clusters within the radiation of USCα PmoA clade, it does not correspond to the major group of PmoA sequences, which are most often retrieved from acidic upland soils. The exact diversity range of USCα methanotrophs, therefore, remains unclear.
This study was undertaken in order to extend the currently available information about the phylogenetic diversity of USCα methanotrophs using unique samples of Russian tundra upland soil, where USCα represents the only detectable methanotroph group.
2. Materials and Methods
2.1. Study Site
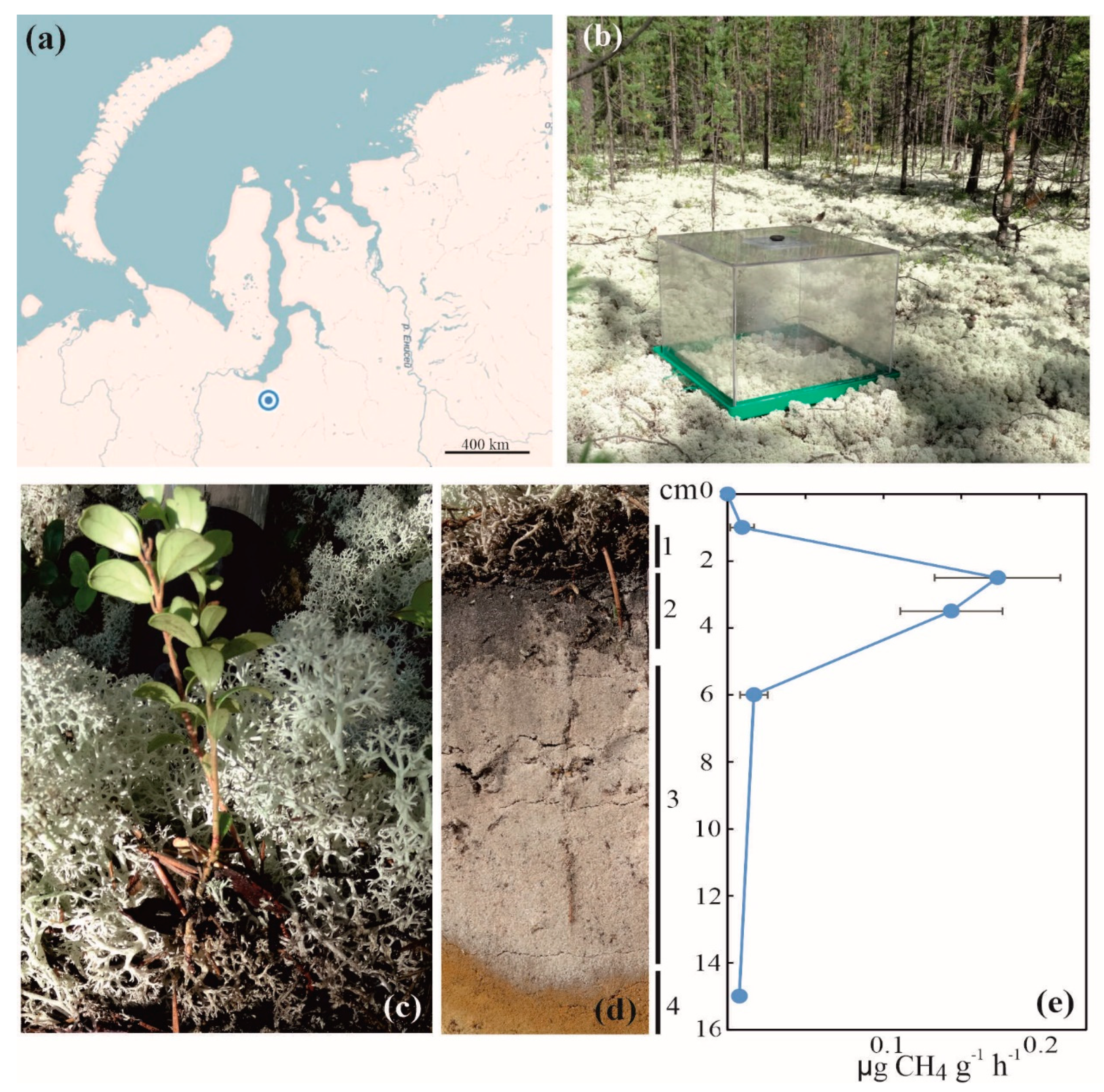
Field studies were carried out in July 2014 in the lichen-dominated pine (Pinus sibirica) forest within a permafrost-free zone of tundra near the Nadym town, the Yamalo-Nenetsky Autonomy, North Siberia (65°36′07.1′′ N, 72°44′ 39.5′′ E) (Figure 1a). The soil vegetation cover was composed of Cladonia and Cetraria species; Vaccinium spp., Ledum palustre, and Polytrichum commune were also present (Figure 1b,c). Analysis of the soil profile revealed the presence of litter, thin organic layer, gray sand, and sandy subsoil (Figure 1d). A set of individual soil samples (~500 g each) was collected over the soil profiles of three sampling plots located on a distance of 20–30 m from each other. The collected soil was transported to the laboratory, homogenized, and frozen for further molecular analyses within 1 day after sampling.
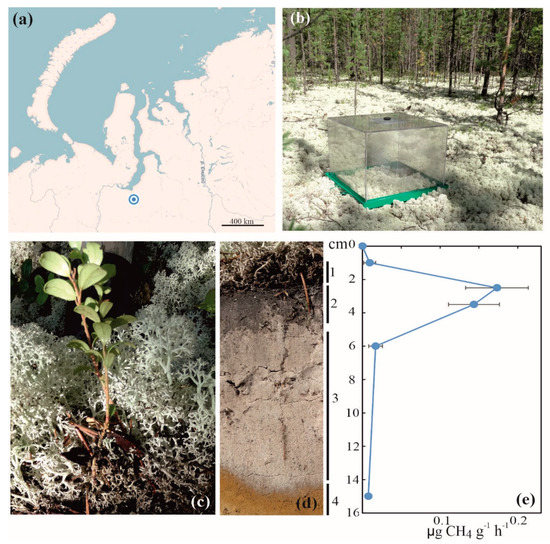
Figure 1.
Location and some characteristics of the study site: (a) location of the study site on the map of northern Russia; (b) CH4 flux measurements by static chambers; (c) vegetation cover at the study site; (d) soil profile: 1—litter, 2—organic layer, 3—gray sand, and 4—sandy subsoil; (e) distribution of CH4 oxidation activity over the soil profile.
2.2. Methane Flux Measurements
Methane fluxes were measured by the static chamber method [29,30]. Concentration of CH4 in the samples was determined using a Kristall-5000 chromatograph (Khromatek, Yoshkar-Ola, Russia) with a flame ionization detector. Methane flux was calculated in mg CH4-C m−2 day−1.
2.3. Determination of CH4 Oxidation Activity of Soil Samples
Methane oxidation rates were determined as described in our previous study [31]. Briefly, weighted portions of wet soil (10 g) sampled from different layers were placed in sterile glass vials 160 mL in volume, which were then sealed hermetically. Methane was injected in the flasks up to the concentration of about 1000 ppm. The vials were incubated at room temperature for 48 h. Samples of the gas phase (0.5 mL) were taken from the flasks periodically and analyzed for methane concentration on a Kristall 5000 chromatograph (Khromatek, Russia). The rate of methane oxidation by the soil samples was calculated in μg CH4 g−1 of wet soil h−1.
2.4. Chemical Analysis of the Soil Samples
The analysis was carried out by certified standard techniques in the analytical center of the MSU Soil Science faculty. The total contents of organic C and N in the soil were measured using the vario MACRO Cube analyzer (Elementar Analysensysteme GmbH, Germany). Sulfate concentrations were measured using the Dionex ICS-2000 Ion Chromatography System (Dionex, Sunnyvale, CA, USA). The contents of Ca, Mg, P, and Fe were determined by means of inductively coupled plasma mass spectrometry (ICP-MS Agilent 7500a, Agilent, Santa Clara, CA, USA).
2.5. DNA Extraction, PCR Amplification, and Illumina Sequencing
Since highest methane oxidation rates were detected in soil collected from the surface organic layer at a depth of 2–5 cm (see Results), these samples were used for molecular diversity studies. Total DNA extracts were obtained from three individual soil samples (~0.5 g wet weight) collected from the three sampling plots. The procedure used for DNA extraction and purification is described in our earlier published study [32]. Three DNA extracts were obtained from each soil sample and pooled together for further use in PCR. Fragments of 16S rRNA gene corresponding to the V4 region were PCR-amplified from the DNA samples, purified, and sequenced using the 300PE protocol on MiSeq System (Illumina, USA) as outlined elsewhere [32]. The obtained reads were quality filtered and trimmed using CLC Genomics Workbench 7.5 (Qiagen, Germany), after which overlapping paired-end reads were merged with SeqPrep tool (https://github.com/jstjohn/SeqPrep).
2.6. Bioinformatic Analysis of Amplicon Sequences
The 16S rRNA gene reads obtained from the studied soil samples were analyzed with QIIME 2 v.2018.8 (https://qiime2.org) [33]. Sequence quality control, denoising, and chimera filtering were performed by using DADA2 plugin [34]. The reads were clustered into operational taxonomic units (OTUs) by using VSEARCH plugin [35] with open-reference function and Silva v. 132 database [36,37] with 97% identity.
2.7. Molecular Identification of Methanotrophic Bacteria
Total DNA extracted from the soil was used as a template in PCR with the primers A189f (GGNGACTGGGACTTCTTG) and A682r (GAASGCNGAGAAGAASGC) specific to the pmoA gene coding for the active-site polypeptide of particulate methane monooxygenase (pMMO) [38]. DNA from the methanotrophic bacterium Methylocystis heyeri H2T was used as the positive control. PCR was performed in a PE GeneAmp PCR System 9700 (Perkin-Elmer Applied Biosystems, United States) thermocycler. The reaction mixture contained 0.5–1.0 μL DNA, A189f and A682r primers, 1 μL each, 50 μL MasterMix (Promega), and sterile water to the total volume of 100 μL. The thermal profile of the reaction was as follows: initial denaturation (1 min at 94 °C); 33 cycles of denaturation (1 min at 94 °C), primer annealing (1 min at 55 °C), and elongation (1 min at 72 °C), followed by final elongation step (7 min at 72 °C). PCR products were examined by electrophoresis in 1.2% agarose gel followed by ethidium bromide staining and visualization of reaction products under a UV transilluminator. Products of three independent reactions obtained with the template DNA of three extracts were pooled together and used for further cloning. Cloning was performed using the pGem-T Easy Vector System II (Promega) according to the manufacturer’s recommendations. Recombinant clones were selected using the blue-white screening and screened for the correct insert size (approximately 530 bp) with vector-specific primers T7 and Sp6 (Promega). Plasmid DNA was purified using a Wizard Plus Minipreps DNA Purification System (Promega). Nucleotide sequences of the cloned DNA fragments were determined on an ABI 377A (Perkin-Elmer Applied Biosystems) sequencer. Phylogenetic trees were constructed using the ARB software package [39].
2.8. qPCR-based Quantification of Methanotrophs
Quantification of pmoA genes was performed according to a previously described method [40] on a the StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, USA) using qPCRmix-HS SYBR Kit (Evrogen, Russia). Concentration of pmoA genes was estimated using the primer system highly specific to the pmoA sequences retrieved from the examined soil samples at the previous stage. A primer system was designed using Primrose software [41]: pmoA-Cl-F (5′-GCA ATA TGG CAC GCT GAT GT-3′) and pmoA-Cl-R (5′-ATG TAT TCG GGC ATC GAG GTA-3′). qPCR thermal program was adjusted experimentally according to the standard protocol: 94 °C—20”; 62 °C—20”; 72 °C—15”; primers conc.—0.5 μM; Mg++ conc.—3 mM. One of the pmoA amplicons inserted in the pGEM®-T vector was used to generate a standard curve. Standards, samples and negative controls were run in triplicate. The correlation coefficient for standard curve was 0.997.
2.9. Sequence Accession Numbers
The 16S rRNA gene reads retrieved using Illumina pair-end sequencing from the soil (raw data) have been deposited under the BioProject number PRJNA344855 in the NCBI Sequence Read Archive, with the accession numbers SRX2207619-SRX2207621. Nucleotide sequences of the pmoA gene fragments determined in this study were deposited in GenBank under accession nos. KX534007–KX534056. Identical nucleotide sequences were not deposited.
3. Results and Discussion
3.1. Measurements of CH4 Fluxes
Measurements of in situ surface CH4 fluxes at the studied forested tundra site in July 2014 showed atmospheric CH4 uptake with rates −0.4 to −0.6 mg CH4-C m−2 day−1. These CH4 uptake rates are comparable with those measured at the McGill Arctic Research Station at Expedition Fjord, AHI, Canada (−0.1 to −0.8 mg CH4-C m−2 day−1) [24], Quttinirpaaq National Park, Canada (−1.37 ± 0.06 mg CH4 m−2 day−1) [42], and other terrestrial systems at lower latitudes (−0.1 to −1.0 mg CH4-C m−2 day−1) [43].
3.2. Distribution of Methane Oxidation Activity over the Soil Profile
Soil profiles in the three sampling plots were highly similar to each other and included litter, thin dark-colored organic layer, gray sand, and sandy subsoil (see 1–4 in Figure 1d). The surface organic layer and the underlying sandy soil displayed a number of contrasting characteristics (Table 1). The former had a pH 4.1, while the latter was nearly neutral (pH 6.1). The content of organic carbon in the surface dark-colored layer was one order of magnitude higher than that in the sandy soil (Table 1). The maximal methane oxidation rates, 0.15–0.18 µg CH4 g−1 h−1, were detected in samples taken from the organic soil layer, while CH4 oxidation activity in the sandy soil layer was nearly non-measurable (Figure 1e). Further molecular analyses, therefore, were performed with DNA extracts obtained from the surface organic soil layer.

Table 1.
Chemical composition of soil samples.
3.3. Identification and Quantification of Methanotrophs Using pmoA-Based Analysis
A total of 222 pmoA gene clone sequences were retrieved from the studied DNA extracts. Of these, only 50 nucleotide sequences were unique and were deposited in the GenBank. These 50 sequences represented 7 closely related pmoA phylotypes grouped on the basis of 98% nucleotide sequence identity (Table 2).

Table 2.
The phylotypes of pmoA gene sequences retrieved from lichen-dominated tundra soil.
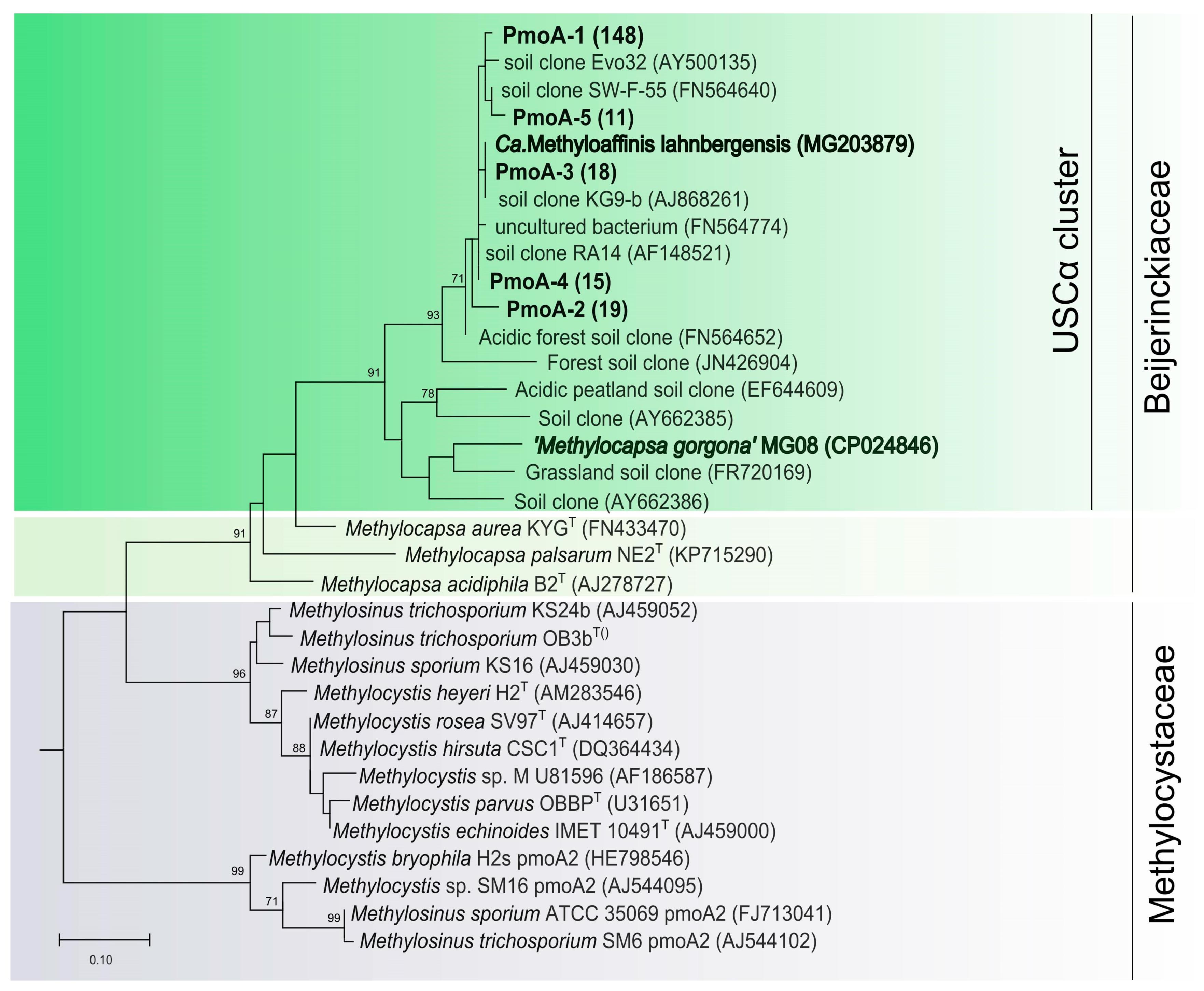
All of them belonged to Beijerinckiaceae methanotrophs and affiliated with the USCα clade (Figure 2). The most abundant phylotype, pmoA-1, was represented by 148 sequences and displayed 97.5% nucleotide identity to pmoA of Candidatus Methyloaffinis lahnbergensis. If grouped on the basis of 93% nucleotide sequence identity as recommended by Degelmann et al. (2010) [21], all pmoA sequences obtained in our study belong to one species-level OTU, thus suggesting extremely low methanotroph diversity in the tundra soil.
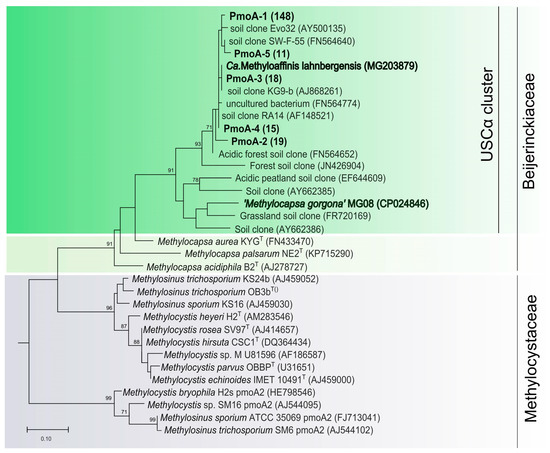
Figure 2.
Phylogenetic tree constructed based on 201 amino acid sites of partial PmoA sequences obtained from a forested tundra soil (shown in bold), as well as PmoA from representative members of the families Beijerinckiaceae and Methylocystaceae and some environmental clone sequences. The scale bar corresponds to 0.1 substitutions per amino acid position.
The pool of pmoA sequences retrieved in this study was used to design the primers for specific detection of the target methanotroph group (see Methods). The qPCR-detected pmoA gene copy numbers were highly similar in all three sampling plots and were within a range of 5.9–8.3 × 106 copies g−1 of wet soil or 6.6–9.7 × 106 copies g−1 of dry soil (given water content of 10.7–14.7%). These values fall within the range of USCα pmoA gene numbers reported earlier for various soils exhibiting atmospheric CH4 consumption, 0.9 × 106–1.2 × 108 copies per gram of dry soil [7,21,44,45]. Highest USCα pmoA gene numbers, 0.3–1.2 × 108 copies per gram of dry soil, have so far been reported for boreal forest soils in Germany [21].
3.4. Molecular Analysis of Bacterial Diversity in Tundra Soil
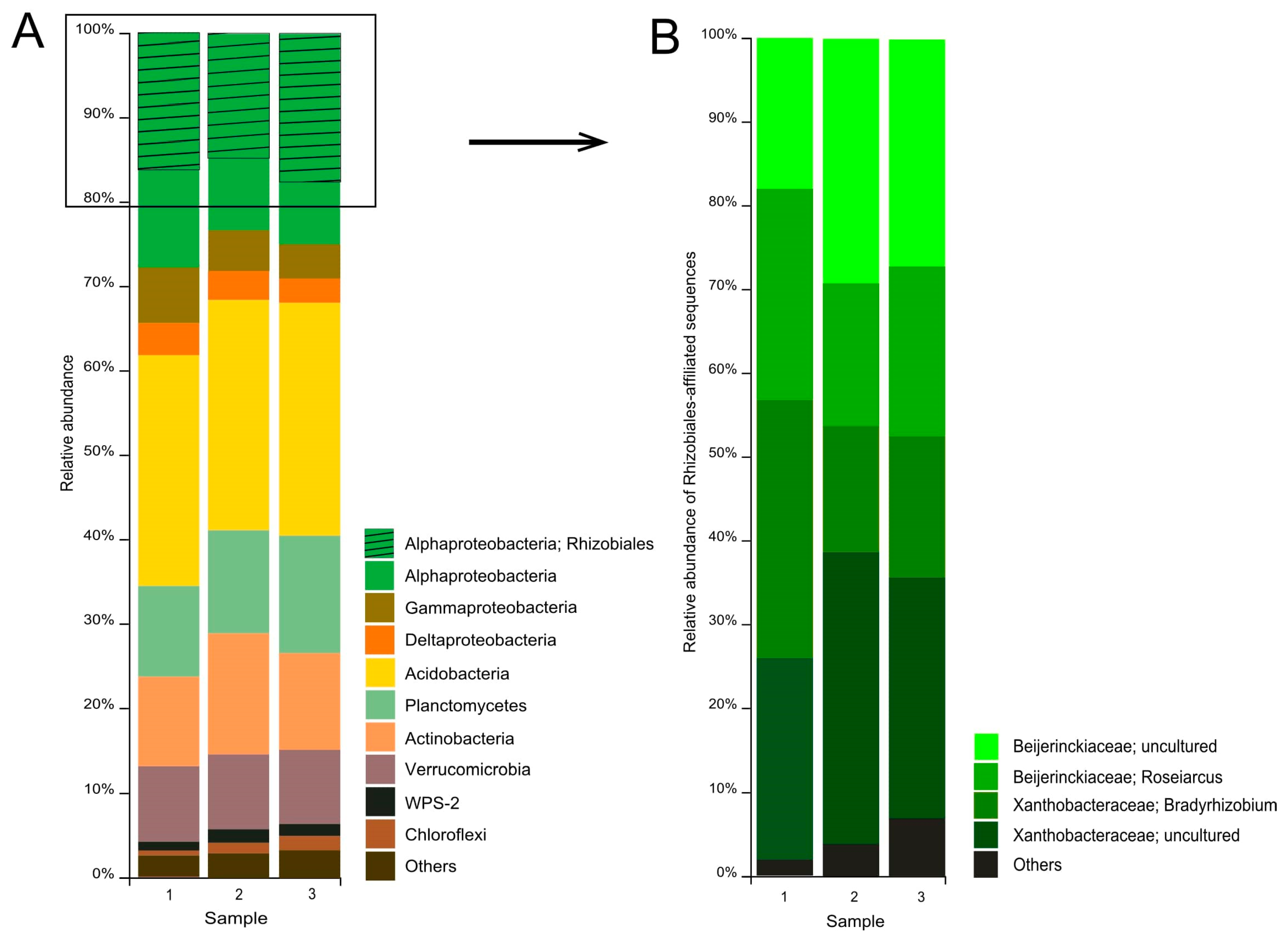
A total of 712,393 partial 16S rRNA gene sequences (mean amplicon length 253 bp) were retrieved from the examined tundra soil. Of these, 496,695 reads were retained after quality filtering, denoising, and removing chimeras. Predominant prokaryote groups in the communities from three sites were the Acidobacteria (27.5 ± 0.1% of the total number of 16S rRNA gene fragments, mean ± SE), Alphaproteobacteria (25.2 ± 1.0%), Planctomycetes (12.3 ± 0.8%), Actinobacteria (12.1 ± 0.9%), Verrucomicrobia (9.3 ± 0.1%), Gammaproteobacteria (5.3 ± 0.7%), and Deltaproteobacteria (3.4 ± 0.2%) (Figure 3A). Minor bacterial groups included WPS-2, Chloroflexi, Cyanobacteria, Firmicutes, and others.
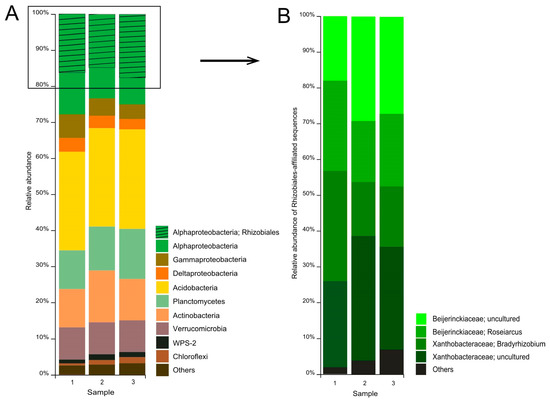
Figure 3.
Bacteria community composition in forested tundra soil according to the results of Illumina-based sequencing of 16S rRNA genes. The composition is displayed at the (A) phylum level and (B) family level of the alphaproteobacterial order Rhizobiales.
The largest group among the Alphaproteobacteria was the order Rhizobiales (64 ± 0.7% of the number of 16S rRNA gene fragments affiliated with the Alphaproteobacteria). Two groups of Rhizobiales-affiliated sequences were represented by reads taxonomically classified within the genera Roseiarcus (20.8 ± 1.9% of all Rhizobiales-affiliated reads) and Bradyrhizobium (20.9 ± 4.1%) (Figure 3B).
The third group of Rhizobiales-affiliated sequences belonged to uncultured members of the Xanthobacteraceae (29.2 ± 2.5%). Finally, the fourth group of sequences (25.8 ± 2.8%) was identified as belonging to uncultured Beijerinckiaceae. Further analysis of these sequences revealed that they are represented by 8 phylotypes (Table 3).

Table 3.
Major operational taxonomic units (OTUs) of 16S rRNA gene sequences affiliated with the family Beijerinckiaceae.
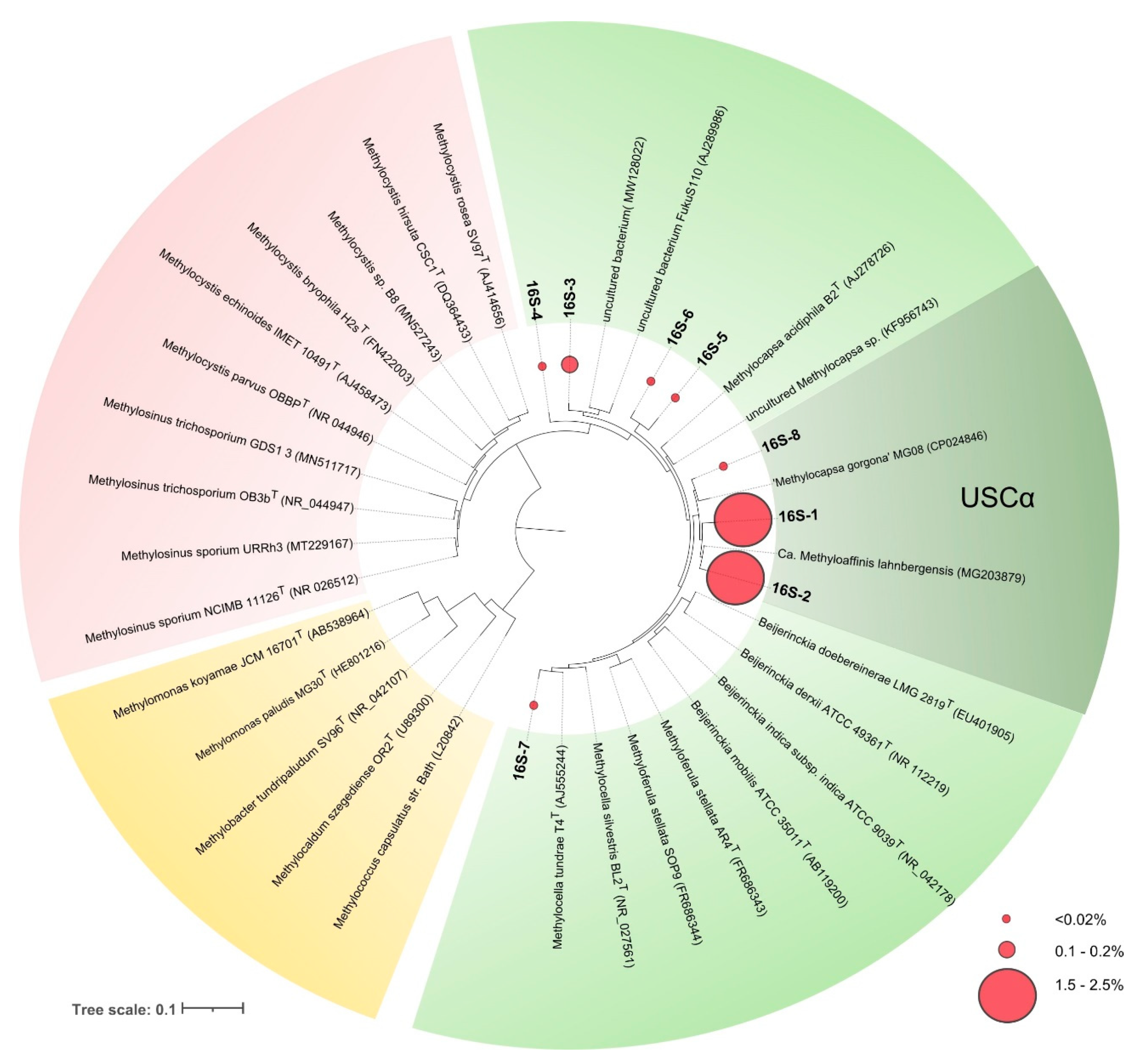
Two of these phylotypes, 16S-1 and 16S-2, were most abundant and were represented by 11,712 and 9446 reads, comprising 2.4 and 1.9% of all bacterial sequences retrieved from the studied soil. Phylotypes 16S-1 and 16S-2 displayed 99 and 100% sequence similarity to 16S rRNA gene sequence of Candidatus Methyloaffinis lahnbergensis and 98.6 and 97.6% sequence similarity to 16S rRNA gene sequence of “Methylocapsa gorgona” MG08, respectively (Table 3, Figure 4).
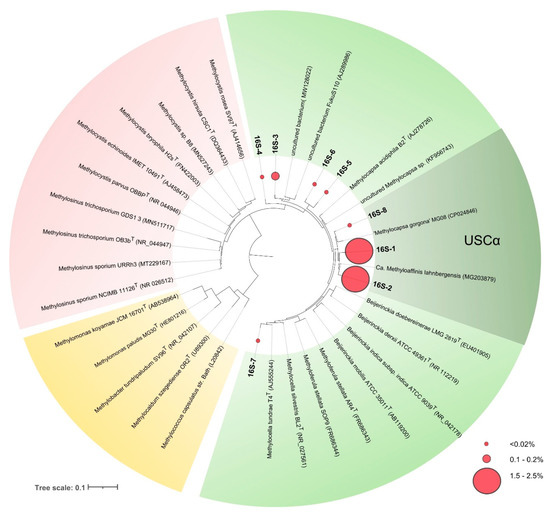
Figure 4.
16S rRNA gene-based maximum-likelihood tree showing the phylogenetic position of sequences retrieved from acidic tundra soil in relation to Candidatus Methyloaffinis lahnbergensis, “Methylocapsa gorgona” MG08 and some representatives of the family Beijerinckiaceae (shown in green), Methylocystaceae (red), and Methylococcaceae (yellow). Red circles of different sizes show relative abundances of the corresponding OTUs. The scale bar corresponds to 0.1 substitutions per nucleotide position.
Notably, no 16S rRNA gene fragments from well-studied methanotrophs of the order Methylococcales and the family Methylocystaceae were detected within the pool of reads retrieved in our study, which fully corresponds to the results of pmoA-based diversity analysis.
The results obtained in our study, therefore, characterize subarctic sandy upland soils as a unique habitat colonized mostly or exclusively by USCα methanotrophs. As shown by the recent 16S rRNA gene-based environmental distribution survey of USCα methanotrophs [27], arctic and subarctic soils are among the major habitats of these bacteria. The ecological advantage of USCα members over other methanotrophs, most likely, is explained by the fact that these soils, in contrast to arctic wetlands and boreal upland soils, are exposed to atmospheric methane concentrations only. While arctic wetlands may emit methane, arctic desert soils function as a constant sink for atmospheric CH4 [42]. One of our previous studies on methanotroph diversity was performed in a subarctic wetland with mosaic cover of Sphagnum mosses and lichens [31]. This wetland is in a close proximity to the study site examined in this work (a distance of 14.5 km). While USCα methanotrophs were also detected in that subarctic wetland, the indigenous methanotroph community was by far more diverse and included conventional type I and type II methanotrophs as well [31]. Interestingly, members of the genera Methylocystis and Methylosinus, which contain Pmo2 enzyme catalyzing oxidation of CH4 at atmospheric levels [46,47,48] and are commonly detected in upland boreal soils [5,7,18,21], were absent from the studied tundra soil. As suggested earlier, these methanotrophs cannot sustain activity and growth on atmospheric methane alone, and their long-term survival depends on availability of methane produced in anaerobic micro-sites of upland soils [5]. Such an in situ methane production does not occur in well-aerated sandy soils examined in this study and, therefore, Pmo2-possessing Methylocystis and Methylosinus species cannot compete with USCα methanotrophs for colonizing this niche.
As seen from Figure 2, most methanotrophs detected in lichen-dominated tundra soil were closely related to Candidatus Methyloaffinis lahnbergensis [27] and not to “Methylocapsa gorgona” MG08 [28]. Although phylogenetically close, these methanotrophs may possess some differences in physiology and adaptation potential, which are reflected in their distribution pattern. Isolation of Candidatus Methyloaffinis-like bacteria remains one of the key challenges in methanotroph cultivation and subarctic sandy soils are one of the promising sources for these isolation studies.
In summary, our study showed that upland soils of a lichen-dominated forested tundra function as a sink for atmospheric CH4. Methane-oxidizing communities in these soils are composed exclusively of high-affinity USCα methanotrophs, which are localized in a very thin organic surface soil layer underlying the lichen cover. Possible disturbances of this surface soil layer due to anthropogenic activities may result in degradation of this fragile methane-oxidizing microbial filter and loss of its function.
Author Contributions
Conceptualization, S.N.D.; investigation, S.E.B., O.V.D., and A.Y.M.; data curation, A.A.I. and A.Y.M.; writing—original draft preparation, S.E.B., A.Y.M., and S.N.D.; writing—review and editing, S.N.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Higher Education of the Russian Federation.
Acknowledgments
The authors are grateful to Irina Gagarinova (Center for Arctic Research, Nadym) for her help in the field.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dlugokencky, E.J.; Nisbet, E.G.; Fisher, R.; Lowry, D. Global atmospheric methane: Budget, changes and dangers. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 2058–2072. [Google Scholar] [CrossRef] [PubMed]
- Kirschke, S.; Bousquet, P.; Ciais, P.; Saunois, M.; Canadell, J.G.; Dlugokencky, E.J.; Bergamaschi, P.; Bergmann, D.; Blake, D.R.; Bruhwiler, L.; et al. Three decades of global methane sources and sinks. Nat. Geosci. 2013, 6, 813–823. [Google Scholar] [CrossRef]
- Ciais, P.; Sabine, C.; Bala, G.; Bopp, L.; Brovkin, V.; Canadell, J.; Chhabra, A.; DeFries, R.; Galloway, J.; Heimann, M.; et al. Carbon and other biogeochemical cycles. In Climate Change 2013 the Physical Science Basis: Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, UK, 2013; pp. 465–570. [Google Scholar]
- Prather, M.J.; Holmes, C.D. Overexplaining or underexplaining methane’s role in climate change. Proc. Natl. Acad. Sci. USA 2017, 114, 5324–5326. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F. The soil methane sink. In Greenhouse Gas Sinks; Reay, D.S., Hewitt, C.N., Smith, K.A., Grace, J., Eds.; CABI: Wallingford, UK, 2007; pp. 152–170. [Google Scholar]
- Shallcross, D.E.; Butenhoff, C. The atmospheric methane sink. In Greenhouse Gas Sinks; Reay, D.S., Hewitt, C.N., Smith, K.A., Grace, J., Eds.; CABI: Wallingford, UK, 2007; pp. 171–184. [Google Scholar]
- Kolb, S. The quest for atmospheric methane oxidizers in forest soils. Environ. Microbiol. Rep. 2009, 1, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Rosenberg, E., Delong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; pp. 267–285. [Google Scholar]
- Khmelenina, V.N.; Colin Murrell, J.; Smith, T.J.; Trotsenko, Y.A. Physiology and biochemistry of the aerobic methanotrophs. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Rojo, F., Ed.; Springer: Cham, Switzerland, 2019; pp. 1–25. [Google Scholar]
- Dedysh, S.N.; Knief, C. Diversity and phylogeny of described aerobic methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer: Cham, Switzerland, 2018; pp. 17–42. [Google Scholar]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Bender, M.; Conrad, R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Lett. 1992, 101, 261–270. [Google Scholar] [CrossRef]
- Holmes, A.J.; Roslev, P.; McDonald, I.R.; Iversen, N.; Henriksen, K.; Murrell, J.C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 1999, 65, 3312–3318. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Horz, H.P.; Dunfield, P.F.; Liesack, W. A novel pmoA lineage represented by the acidophilic methanotrophic bacterium Methylocapsa acidiphila [correction of acidophila] B2. Arch. Microbiol. 2001, 177, 117–121. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Semrau, J.D.; Liesack, W.; Tiedje, J.M. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 2002, 52, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Knief, C.; Lipski, A.; Dunfield, P.F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Henckel, T.; Jäckel, U.; Schnell, S.; Conrad, R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 2000, 66, 1801–1808. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Holmes, A.J.; Olsen, R.A.; Murrell, J.C. Detection of methane oxidizing bacteria in forest soil by monooxygenase PCR amplification. Microb. Ecol. 2000, 39, 282–289. [Google Scholar] [PubMed]
- Degelmann, D.M.; Borken, W.; Drake, H.L.; Kolb, S. Different atmospheric methane-oxidizing communities in european beech and norway spruce soils. Appl. Environ. Microbiol. 2010, 10, 3228–3235. [Google Scholar] [CrossRef]
- Dörr, N.; Glaser, B.; Kolb, S. Methanotrophic communities in brazilian ferralsols from naturally forested, afforested, and agricultural sites. Appl. Environ. Microbiol. 2010, 76, 1307–1310. [Google Scholar] [CrossRef]
- Martineau, C.; Pan, Y.; Bodrossy, L.; Yergeau, E.; Whyte, L.G.; Greer, C.W. Atmospheric methane oxidizers are present and active in Canadian high Arctic soils. FEMS Microbiol. Ecol. 2014, 89, 257–269. [Google Scholar] [CrossRef]
- Lau, M.C.Y.; Stackhouse, B.T.; Layton, A.C.; Chauhan, A.; Vishnivetskaya, T.A.; Chourey, K.; Ronholm, J.; Mykytczuk, N.C.S.; Bennett, P.C.; Lamarche-Gagnon, G.; et al. An active atmospheric methane sink in high Arctic mineral cryosols. ISME J. 2015, 9, 1880–1891. [Google Scholar] [CrossRef]
- Ricke, P.; Kolb, S.; Braker, G. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 2005, 71, 1671–1673. [Google Scholar] [CrossRef]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R.; et al. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 12, 2544–2558. [Google Scholar] [CrossRef]
- Pratscher, J.; Vollmers, J.; Wiegand, S.; Dumont, M.G.; Kaster, A.K. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing upland soil cluster α. Environ. Microbiol. 2018, 20, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Tveit, A.T.; Hestnes, A.G.; Robinson, S.L.; Schintlmeister, A.; Dedysh, S.N.; Jehmlich, N.; von Bergen, M.; Herbold, C.; Wagner, M.; Richter, A.; et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc. Natl. Acad. Sci. USA 2019, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Glagolev, M.; Kleptsova, I.; Filippov, I.; Maksyutov, S.; MacHida, T. Regional methane emission from West Siberia mire landscapes. Environ. Res. Lett. 2011, 6, 045214. [Google Scholar] [CrossRef]
- Glagolev, M.V.; Sabrekov, A.F.; Kleptsova, I.E.; Filippov, I.V.; Lapshina, E.D.; Machida, T.; Maksyutov, S.S. Methane emission from bogs in the subtaiga of Western Siberia: The development of standard model. Eurasian Soil Sci. 2012, 45, 947–957. [Google Scholar] [CrossRef]
- Danilova, O.V.; Belova, S.E.; Gagarinova, I.V.; Dedysh, S.N. Microbial community composition and methanotroph diversity of a subarctic wetland in Russia. Microbiology 2016, 85, 583–591. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Kulichevskaya, I.S.; Merkel, A.Y.; Toshchakov, S.V.; Dedysh, S.N. High diversity of Planctomycetes in soils of two lichen-dominated sub-arctic ecosystems of northwestern Siberia. Front. Microbiol. 2016, 7, 2065. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar, A.; Buchner, A.; Lai, T.; Steppi, S.; Jacob, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 2–3, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Ashelford, K.E.; Weightman, A.J.; Fry, J.C. Primrose: A computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 2002, 30, 3481–3489. [Google Scholar] [CrossRef]
- Emmerton, C.A.; St. Louis, V.L.; Lehnherr, I.; Humphreys, E.R.; Rydz, E.; Kosolofski, H.R. The net exchange of methane with high Arctic landscapes during the summer growing season. Biogeosciences 2014, 11, 3095–3106. [Google Scholar] [CrossRef]
- Luo, G.J.; Kiese, R.; Wolf, B.; Butterbach-Bahl, K. Effects of soil temperature and moisture on methane uptake and nitrous oxide emissions across three different ecosystem types. Biogeosciences 2013, 10, 3205–3219. [Google Scholar] [CrossRef]
- Kolb, S.; Knief, C.; Dunfield, P.F.; Conrad, R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 2005, 7, 1150–1161. [Google Scholar] [CrossRef]
- Pratscher, J.; Dumont, M.G.; Conrad, R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ. Microbiol. 2011, 13, 2692–2701. [Google Scholar] [CrossRef]
- Dunfield, P.F.; Yimga, M. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 2002, 41, 17–26. [Google Scholar] [CrossRef]
- Knief, C.; Dunfield, P.F. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ. Microbiol. 2005, 7, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Baani, M.; Liesack, W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. USA 2008, 105, 10203–10208. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).