Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Physiochemical Analyses
2.3. Sequencing of 16S rRNA Gene Amplicons
2.4. Metagenome Analysis Employing Shotgun Sequencing
2.5. Bioinformatic Analysis
2.5.1. 16S rRNA Gene Amplicon Analysis
2.5.2. Shotgun Metagenome Analysis
2.6. Quantification of ARGs Using qPCR
2.7. Statistical Analyses
3. Results
3.1. Nutrient Analysis of Water Samples
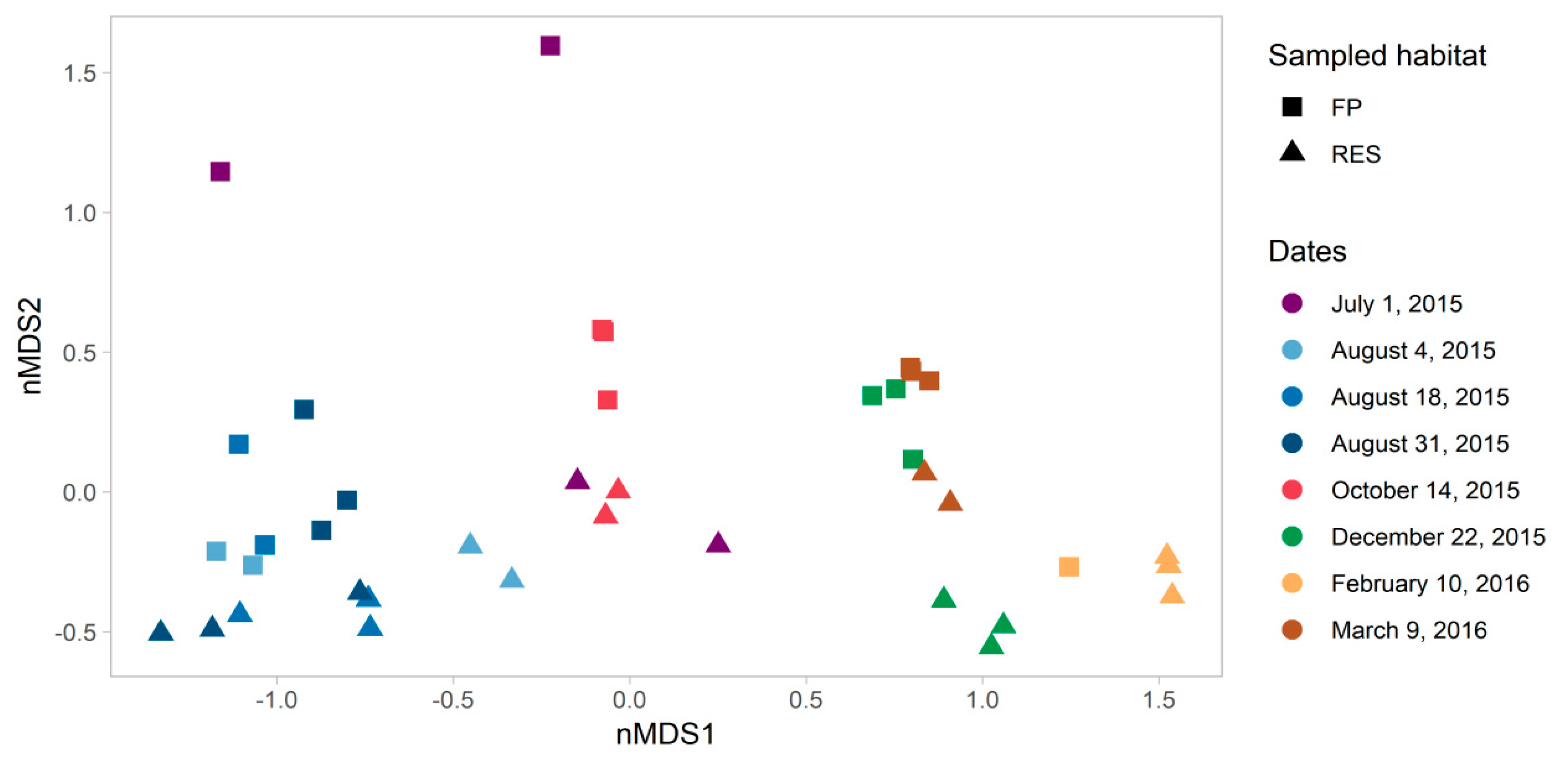
3.2. Microbial Community Analysis
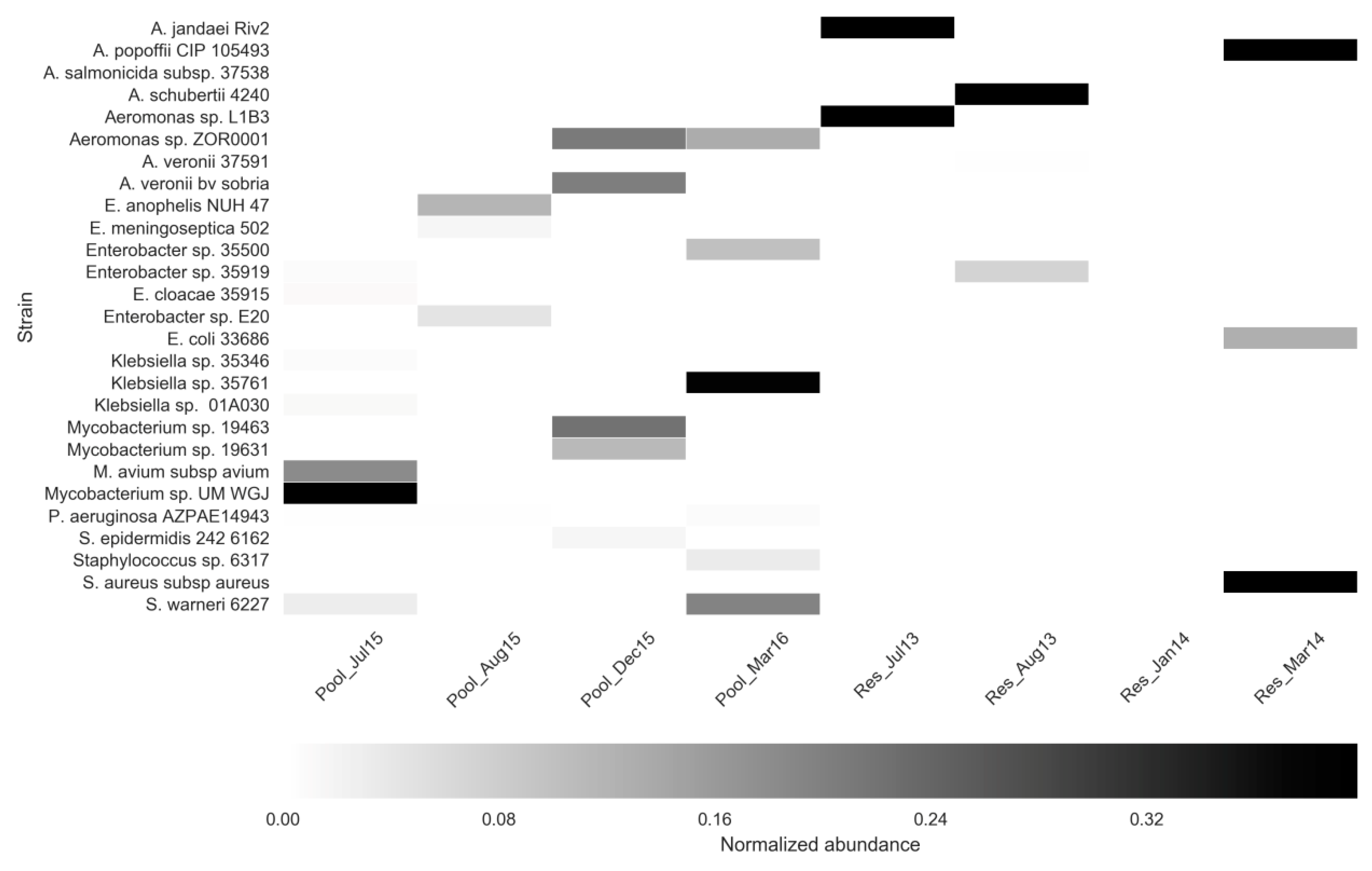
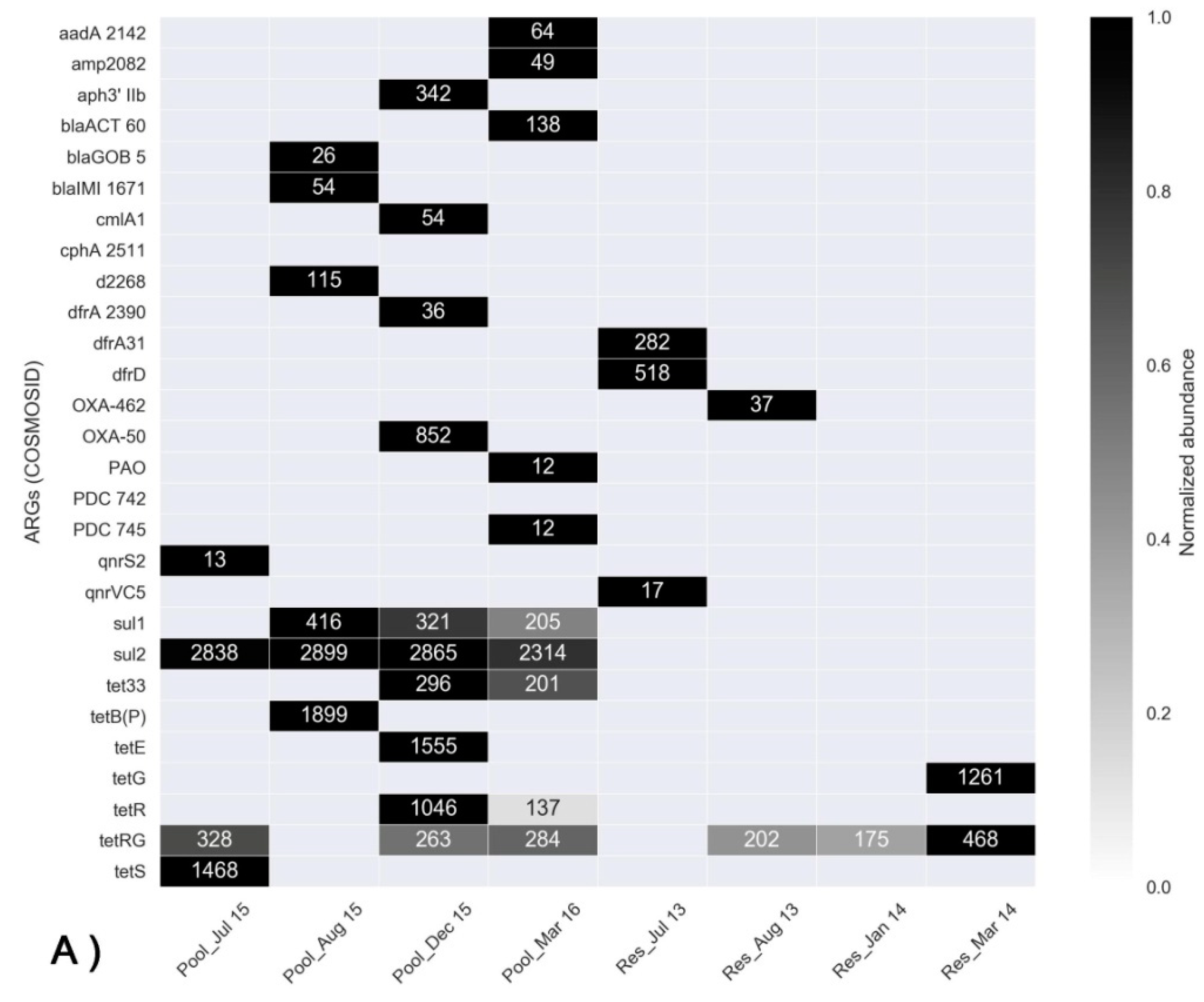
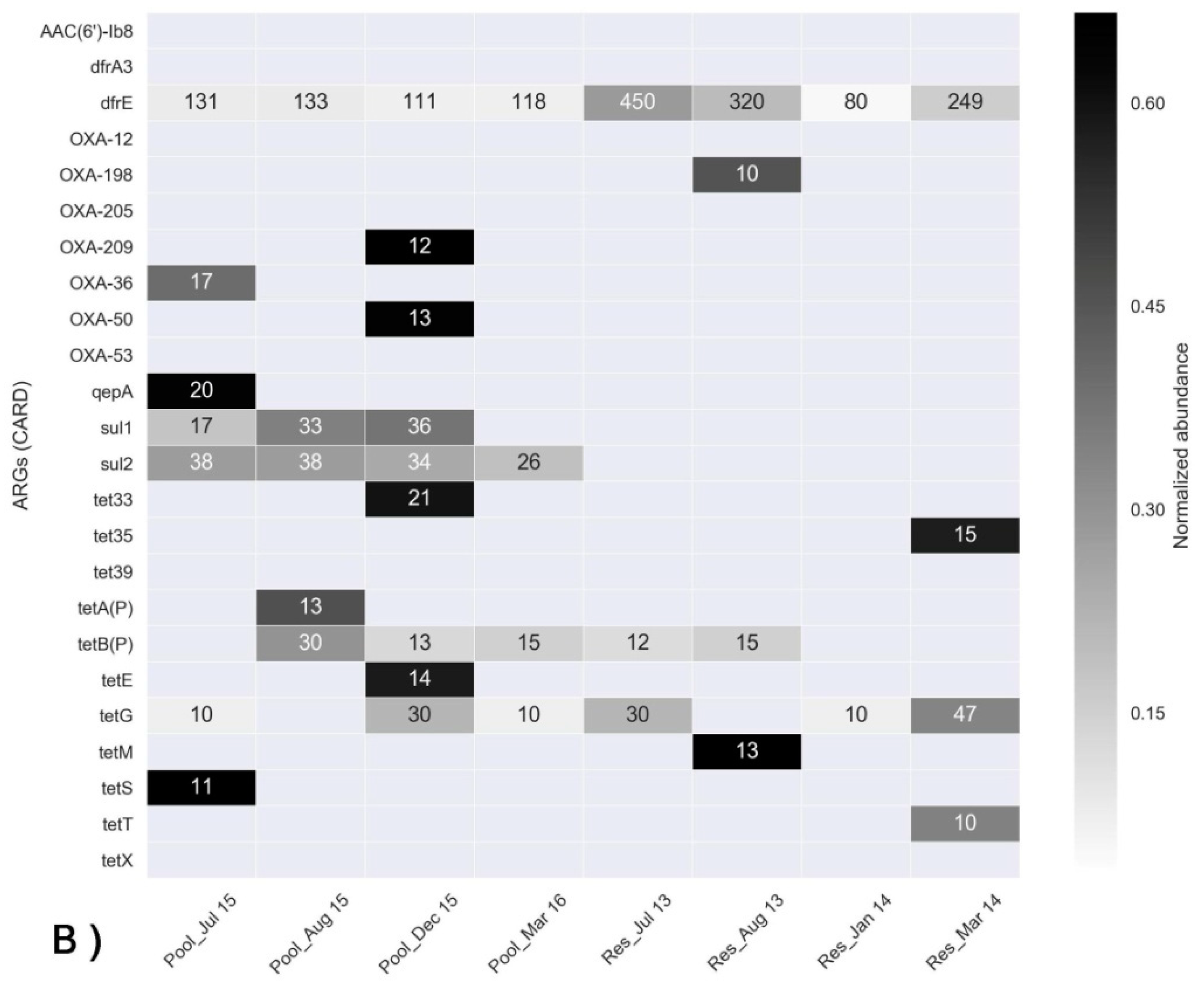
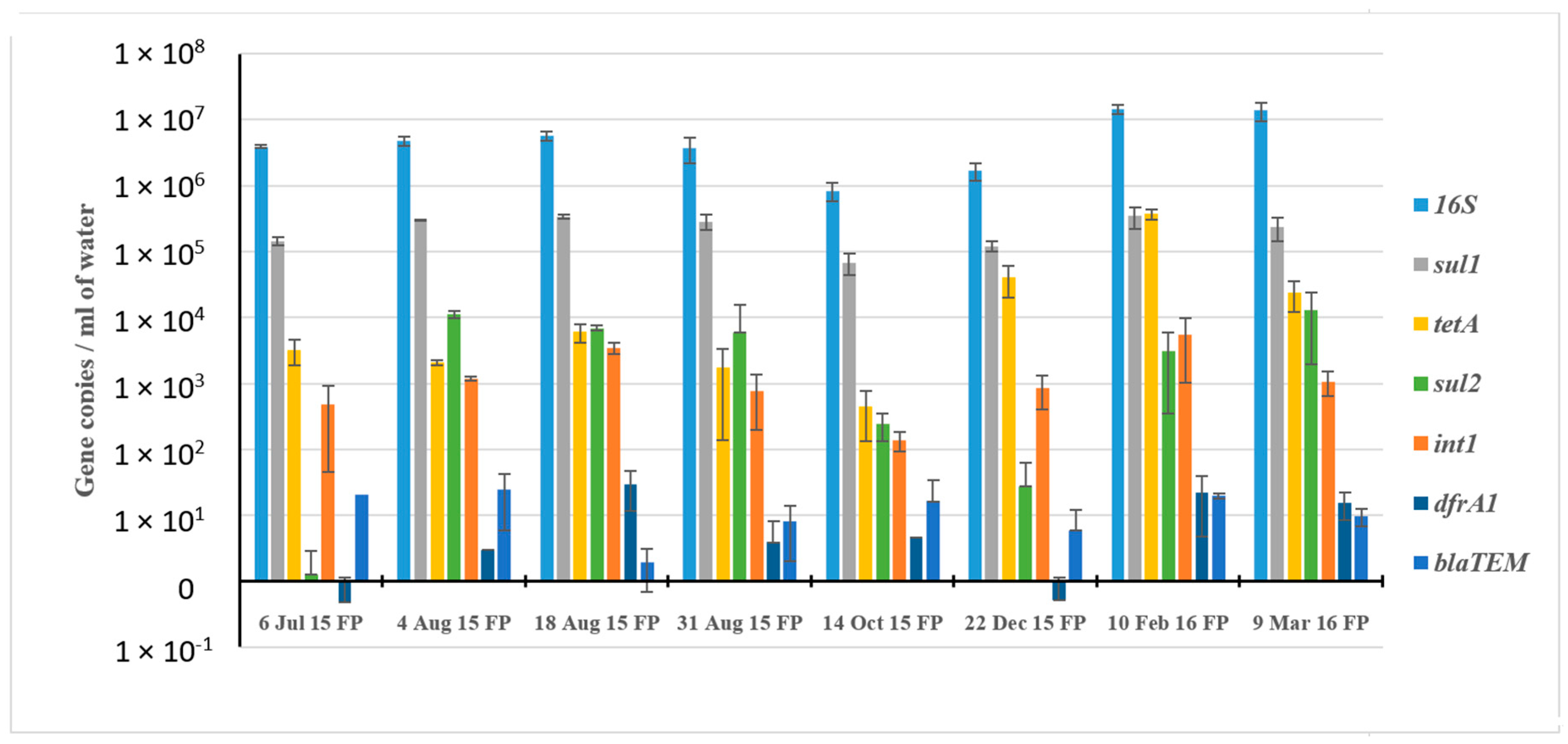
3.3. Assessment of ARGs Using Shotgun Metagenomics and qPCR
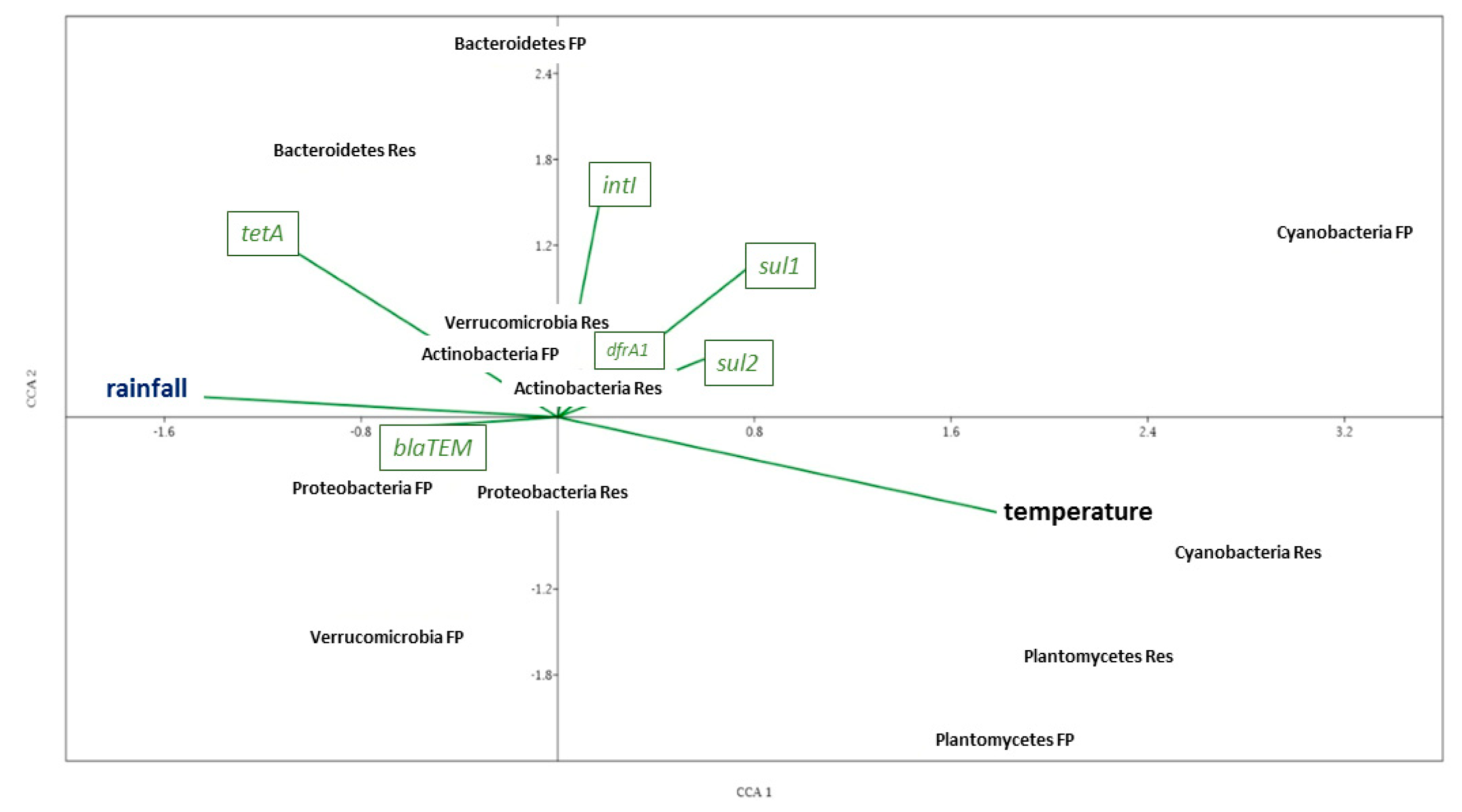
3.4. Correlation Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Zhenling, Z.; Wang, M.; Zeng, Z.-L. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Leeb, M. A shot in the arm. Nature 2004, 431, 892–893. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.Q.A.; Cabello, F.C.; L’Abée-Lund, T.M.; Tomova, A.; Godfrey, H.P.; Buschmann, A.H.; Sørum, H. Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non-aquaculture sites. Environ. Microbiol. 2014, 16, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Genet. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Da Silva, G.J.; Domingues, S. Insights on the Horizontal Gene Transfer of Carbapenemase Determinants in the Opportunistic Pathogen Acinetobacter baumannii. Microorganisms 2016, 4, 29. [Google Scholar] [CrossRef]
- Alderman, D.J.; Hastings, T.S. Antibiotic use in aquaculture: Development of antibiotic resistance—Potential for consumer health risks. Int. J. Food Sci. Technol. 1998, 33, 139–155. [Google Scholar] [CrossRef]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef] [PubMed]
- Patil, H.J.; Benet-Perelberg, A.; Naor, A.; Smirnov, M.; Ofek, T.; Nasser, A.; Minz, D.; Cytryn, E. Evidence of Increased Antibiotic Resistance in Phylogenetically-Diverse Aeromonas Isolates from Semi-Intensive Fish Ponds Treated with Antibiotics. Front. Microbiol. 2016, 7, 1875. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Hoa, T.T.T.; Harada, K.; Warisaya, M.; Asayama, M.; Hinenoya, A.; Lee, J.W.; Phu, T.M.; Ueda, S.; Sumimura, Y.; et al. Water metagenomic analysis reveals low bacterial diversity and the presence of antimicrobial residues and resistance genes in a river containing wastewater from backyard aquacultures in the Mekong Delta, Vietnam. Environ. Pollut. 2017, 222, 294–306. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization: Report of a Joint FAO/OIE/WHO Expert Consultation on Antimicrobial Use in Aquaculture and Antimicrobial Resistance. Seoul, Republic of Korea. 2006. Available online: http://apps.who.int/iris/bitstream/10665/133869/1/9241595124_eng.pdf (accessed on 27 December 2016).
- Tang, Y.; Tao, P.; Tan, J.; Mu, H.; Peng, L.; Yang, D.; Tong, S.; Chen, L. Identification of bacterial community composition in freshwater aquaculture system farming of Litopenaeusvannamei reveals distinct temperature-driven patterns. Int. J. Mol. Sci. 2014, 15, 13663–13680. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Porchas, M.; Vargas-Albores, F. Microbial metagenomics in aquaculture: A potential tool for a deeper insight into the activity. Rev. Aquac. 2017, 9, 42–56. [Google Scholar] [CrossRef]
- Monchy, S.; Sanciu, G.; Jobard, M.; Rasconi, S.; Gerphagnon, M.; Chabé, M.; Cian, A.; Meloni, D.; Niquil, N.; Christaki, U.; et al. Exploring and quantifying fungal diversity in freshwater lake ecosystems using rDNA cloning/sequencing and SSU tag pyrosequencing. Environ. Microbiol. 2011, 13, 1433–1453. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.M.; McHardy, A.C.; Knight, R.T.; Joint, I.; et al. Defining seasonal marine microbial community dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef]
- Quince, C.; Lundin, E.E.; Andreasson, A.N.; Greco, D.; Rafter, J.; Talley, N.J.; Agreus, L.; Andersson, A.F.; Engstrand, L.; D’Amato, M. The impact of Crohn’s disease genes on healthy human gut microbiota: A pilot study. Gut 2013, 62, 952–954. [Google Scholar] [CrossRef]
- Colombo, S.; Arioli, S.; Guglielmetti, S.; Lunelli, F.; Mora, D. Virome-associated antibiotic-resistance genes in an experimental aquaculture facility. FEMS Microbiol. Ecol. 2016, 92, fiw003. [Google Scholar] [CrossRef]
- Kennedy, K.; Hall, M.W.; Lynch, M.D.J.; Moreno-Hagelsieb, G.; Neufeld, J.D. Evaluating Bias of Illumina-Based Bacterial 16S rRNA Gene Profiles. Appl. Environ. Microbiol. 2014, 80, 5717–5722. [Google Scholar] [CrossRef]
- Green, S.; Venkatramanan, R.; Naqib, A. Deconstructing the Polymerase Chain Reaction: Understanding and Correcting Bias Associated with Primer Degeneracies and Primer-Template Mismatches. PLoS ONE 2015, 10, e0128122. [Google Scholar] [CrossRef] [PubMed]
- Bybee, S.M.; Bracken-Grissom, H.; Haynes, B.D.; Hermansen, R.A.; Byers, R.J.; Clement, M.J.; Udall, J.A.; Wilcox, E.R.; Crandall, K.A. Targeted Amplicon Sequencing (TAS): A Scalable Next-Gen Approach to Multilocus, Multitaxa Phylogenetics. Genome Biol. Evol. 2011, 3, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Moonsamy, P.; Williams, T.; Bonella, P.; Holcomb, C.L.; Höglund, B.N.; Hillman, G.; Goodridge, D.; Turenchalk, G.S.; Blake, L.A.; Daigle, D.A.; et al. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array™ system for simplified amplicon library preparation. Tissue Antigens 2013, 81, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Hasan, N.A.; Young, B.A.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; et al. Microbial Community Profiling of Human Saliva Using Shotgun Metagenomic Sequencing. PLoS ONE 2014, 9, e97699. [Google Scholar] [CrossRef]
- Leddy, M.B.; Hasan, N.A.; Subramanian, P.; Heberling, C.; Cotruvo, J.; Colwell, R.R. Characterization of Microbial Signatures From Advanced Treated Wastewater Biofilms. J. Am. Water Work. Assoc. 2017, 109, E503–E512. [Google Scholar] [CrossRef]
- Connelly, S.; Subramanian, P.; Hasan, N.A.; Colwell, R.R.; Kaleko, M. Distinct consequences of amoxicillin and ertapenem exposure in the porcine gut microbiome. Anaerobe 2018, 53, 82–93. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kristiansson, E.; Fick, J.; Janzon, A.; Grabic, R.; Rutgersson, C.; Weijdegård, B.; Söderström, H.; Larsson, D.G.J. Pyrosequencing of Antibiotic-Contaminated River Sediments Reveals High Levels of Resistance and Gene Transfer Elements. PLoS ONE 2011, 6, e17038. [Google Scholar] [CrossRef] [PubMed]
- Bockelmann, U.; Dorries, H.H.; Ayuso-Gabella, M.N.; de Marcay, M.S.; Tandoi, V.; Levantesi, C.; Masciopinto, C.; Van Houtte, E.; Szewzyk, U.; Wintgens, T.; et al. Quantitative PCR Monitoring of Antibiotic Resistance Genes and Bacterial Pathogens in Three European Artificial Groundwater Recharge Systems. Appl. Environ. Microbiol. 2009, 75, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.-C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef]
- Muziasari, W.I.; Managaki, S.; Pärnänen, K.; Karkman, A.; Lyra, C.; Tamminen, M.; Suzuki, S.; Virta, M. Sulfonamide and trimethoprim resistance genes persist in sediments at Baltic Sea aquaculture farms but are not detected in the surrounding environment. PLoS ONE 2014, 9, e92702. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Qiao, M.; Zhang, B.; Cheng, W.-D.; Zhu, Y.-G. Abundance and Diversity of Tetracycline Resistance Genes in Soils Adjacent to Representative Swine Feedlots in China. Environ. Sci. Technol. 2010, 44, 6933–6939. [Google Scholar] [CrossRef] [PubMed]
- Bibbal, D.; Dupouy, V.; Ferré, J.-P.; Toutain, P.-L.; Fayet, O.; Prère, M.-F.; Bousquet-Mélou, A. Impact of Three Ampicillin Dosage Regimens on Selection of Ampicillin Resistance in Enterobacteriaceae and Excretion of blaTEM Genes in Swine Feces. Appl. Environ. Microbiol. 2007, 73, 4785–4790. [Google Scholar] [CrossRef]
- Levesque, C.; Roy, P.H. PCR Analysis of Integrons. In Diagnostic Molecular Microbiology: Principles and Applications; Persing, D.H., Smith, T.F., Tenover, F.C., White, T.J., Eds.; American Society for Microbiology: Washington, DC, USA, 1993; pp. 590–594. [Google Scholar]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Klase, G.; Lee, S.; Liang, S.; Kim, J.; Zo, Y.-G.; Lee, J. The microbiome and antibiotic resistance in integrated fishfarm water: Implications of environmental public health. Sci. Total. Environ. 2019, 649, 1491–1501. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef]
- Ng, C.; Le, T.-H.; Goh, S.G.; Liang, L.; Kim, Y.; Rose, J.B.; Yew-Hoong, K.G. A Comparison of Microbial Water Quality and Diversity for Ballast and Tropical Harbor Waters. PLoS ONE 2015, 10, e0143123. [Google Scholar] [CrossRef]
- Gao, P.; Mao, D.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Suehiro, F.; Tuyen, B.C.; Suzuki, S. Distribution and diversity of tetracycline resistance genes encoding ribosomal protection proteins in Mekong river sediments in Vietnam. FEMS Microbiol. Ecol. 2007, 59, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kobayashi, T.; Suehiro, F.; Tuyen, B.C.; Tana, T.S. High Occurrence Rate of Tetracycline (TC)-Resistant Bacteria and TC Resistance Genes Relates to Microbial Diversity in Sediment of Mekong River Main Waterway. Microbes Environ. 2008, 23, 149–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, H.S.; Lee, J.C.; Kang, H.Y.; Jeong, Y.S.; Lee, E.Y.; Choi, C.H.; Tae, S.H.; Lee, Y.C.; Seol, S.Y.; Cho, D.T. Prevalence of dfr genes associated with integrons and dissemination of dfrA17 among urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 2004, 53, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Gatica, J.; Tripathi, V.; Green, S.; Manaia, C.M.; Berendonk, T.; Cacace, D.; Merlin, C.; Kreuzinger, N.; Schwartz, T.; Fatta-Kassinos, D.; et al. High Throughput Analysis of Integron Gene Cassettes in Wastewater Environments. Environ. Sci. Technol. 2016, 50, 11825–11836. [Google Scholar] [CrossRef] [PubMed]
- Munk, P.; Andersen, V.D.; De Knegt, L.; Jensen, M.S.; Knudsen, B.E.; Lukjancenko, O.; Mordhorst, H.; Clasen, J.; Agersø, Y.; Folkesson, A.; et al. A sampling and metagenomic sequencing-based methodology for monitoring antimicrobial resistance in swine herds. J. Antimicrob. Chemother. 2016, 72, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.C.; Fachmann, M.S.R.; Kiil, K.; Nielsen, E.M.; Hoorfar, J. Gene-Based Pathogen Detection: Can We Use qPCR to Predict the Outcome of Diagnostic Metagenomics? Genes 2017, 8, 332. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, M.; Liu, L.; Liu, X.; Chen, H.; Yang, J. The antibiotic resistome of free-living and particle-attached bacteria under a reservoir cyanobacterial bloom. Environ. Int. 2018, 117, 107–115. [Google Scholar] [CrossRef]
- Fouladkhah, A.; Thompson, B.; Camp, J.S. The Threat of Antibiotic Resistance in Changing Climate. Microorganisms 2020, 8, 748. [Google Scholar] [CrossRef]
- Fukushima, M.; Tomioka, N.; Jutagate, T.; Hiroki, M.; Murata, T.; Preecha, C.; Avakul, P.; Phomikong, P.; Imai, A. The dynamics of pico-sized and bloom-forming cyanobacteria in large water bodies in the Mekong River Basin. PLoS ONE 2017, 12, e0189609. [Google Scholar] [CrossRef]
- Marmen, S.; Blank, L.; Al-Ashhab, A.; Malik, A.; Ganzert, L.; Lalzar, M.; Grossart, H.-P.; Sher, D. The Role of Land Use Types and Water Chemical Properties in Structuring the Microbiomes of a Connected Lake System. bioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Okano, K.; Shimizu, K.; Kawauchi, Y.; Maseda, H.; Utsumi, M.; Zhang, Z.; Neilan, B.A.; Sugiura, N. Characteristics of a Microcystin-Degrading Bacterium under Alkaline Environmental Conditions. J. Toxicol. 2009, 2009, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dziallas, C.; Grossart, H.-P. Temperature and biotic factors influence bacterial communities associated with the cyanobacterium Microcystis sp. Environ. Microbiol. 2011, 13, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Nakagawa, T. Protection against atypical Aeromonas salmonicida infection in carp (Cyprinuscarpio L.) by oral administration of humus extract. J. Vet. Med Sci. 2007, 69, 405–408. [Google Scholar] [CrossRef] [PubMed][Green Version]


| Gene Target | Primer Sequence | Amplicon (bp) | SYBR Green Master Mix Kit (Applied Biosystems) | Reference |
|---|---|---|---|---|
| 16S rRNA (CS1_341F and CS2_806R) | 5′-ACACTGACGACATGGTTCT ACANNNNCCTACGGGAGGCAGCAG-3′ | 195 | FAST | [21] |
| 5′-TACGGTAGCAGAGACTTG GTCTGGACTACHVGGGTWTCTAAT-3′ | ||||
| sul1 | 5′-CGCACCGGAAACATCGCT GCAC-3′ | 163 | FAST | [34] |
| 5′-TGAAGTTCCGCCGCAAGG CTCG-3′ | ||||
| sul2 | 5′-TCCGATGGAGGCCGGTATC TGG-3′ | 191 | POWER | [34] |
| 5′-CGGGAATGCCATCTGCCTT GAG-3′ | ||||
| dfrA1 | 5′-TTCAGGTGGTGGGGAGAT ATAC-3′ | 150 | POWER | [35] |
| 5′-TTAGAGGCGAAGTCTTGG GTAA-3′ | ||||
| tetA | 5′-GCTACATCCTGCTTGCCT TC-3′ | 210 | SELECT | [36] |
| 5′-CATAGATCGCCGTGAAGA GG-3′ | ||||
| blaTEM | 5′-TTCCTGTTTTTGCTCACCC AG-3′ | 113 | SELECT | [37] |
| 5′-CTCAAGGATCTTACCGCTG TTG-3′ | ||||
| intI1 | 5′-CCTCCCGCACGATGATC-3′ | 293 | POWER | [38] |
| 5′-TCCACGCATCGTCAGGC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patil, H.J.; Gatica, J.; Zolti, A.; Benet-Perelberg, A.; Naor, A.; Dror, B.; Al Ashhab, A.; Marman, S.; Hasan, N.A.; Colwell, R.R.; et al. Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment. Microorganisms 2020, 8, 1984. https://doi.org/10.3390/microorganisms8121984
Patil HJ, Gatica J, Zolti A, Benet-Perelberg A, Naor A, Dror B, Al Ashhab A, Marman S, Hasan NA, Colwell RR, et al. Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment. Microorganisms. 2020; 8(12):1984. https://doi.org/10.3390/microorganisms8121984
Chicago/Turabian StylePatil, Hemant J., Joao Gatica, Avihai Zolti, Ayana Benet-Perelberg, Alon Naor, Barak Dror, Ashraf Al Ashhab, Sophi Marman, Nur A. Hasan, Rita R. Colwell, and et al. 2020. "Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment" Microorganisms 8, no. 12: 1984. https://doi.org/10.3390/microorganisms8121984
APA StylePatil, H. J., Gatica, J., Zolti, A., Benet-Perelberg, A., Naor, A., Dror, B., Al Ashhab, A., Marman, S., Hasan, N. A., Colwell, R. R., Sher, D., Minz, D., & Cytryn, E. (2020). Temporal Resistome and Microbial Community Dynamics in an Intensive Aquaculture Facility with Prophylactic Antimicrobial Treatment. Microorganisms, 8(12), 1984. https://doi.org/10.3390/microorganisms8121984







