The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections
Abstract
1. Introduction
2. Materials and Methods
2.1. Amplification of the CTX-M Genes
2.2. Construction of CTX-M-14
- CTXM-14-A55T-F 5′-ACGCCCAGCCGCCCTCCGTGC-3′
- CTXM-14-A55T-R 5′-AGAGATGTGCTGGCCTGGGT-3′
- CTXM-14-P273A C277R-F 5′-CCGCAACAGAACGCAGAGAGCCGCAGAGATGTGCTGGC-3′
- CTXM-14-P273A C277R-R 5′-CTGGGTAAAATAGGTCACCAG-3′
2.3. Recombinant Protein Cloning
- CTX-M-new/14-F: atggcataatggtctcaggccATGGTGACAAAGAGAGTGCAACGG,
- CTX-M-new/14-R: atggcataatggtctcagcgctCAGCCCTTCGGCGATGATTC
- CTX-M-15-F: atggcataatggtctcaggccATGGTTAAAAAATCACTGCGCCAG
- CTX-M-15-R atggcataatggtctcagcgctCAAACCGTCGGTGACGATTTTAG
2.4. Periplasmatic Protein Expression and Purification
2.5. MIC Determination
2.6. In Vitro Hydrolysis Assay
3. Results
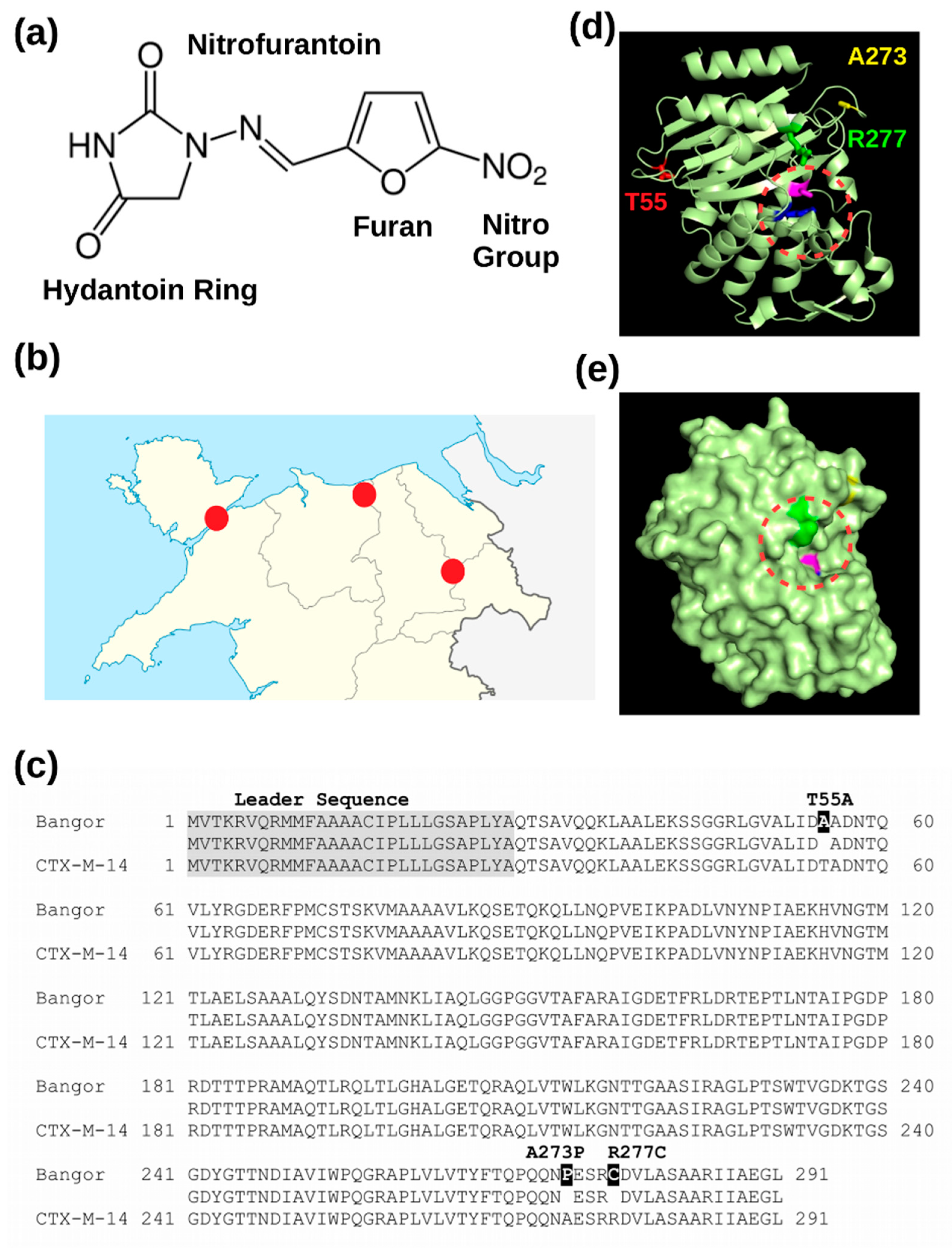
3.1. A Novel Beta-Lactamase Related to CTX-M-14
3.2. The Novel Beta-Lactamase Differs in Three Amino Acid Positions from CTX-M-14
3.3. Recombinant Expression of the New Beta-Lactamase in the Periplasm of E. coli Renders Cells Nitrofurantoin Resistant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Medina, M.; Castillo-Pino, E. An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Tandogdu, Z.; Wagenlehner, F.M.E. Global epidemiology of urinary tract infections. Curr. Opin. Infect. Dis. 2016, 29, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Mandell, G.L.; Bennett, J.E.; Dolin, R. (Eds.) Principles and Practice of Infectious Diseases, 7th ed.; Churchill Livingstone: Philadelphia, PA, USA, 2010; pp. 515–520. [Google Scholar]
- McOsker, C.C.; Fitzpatrick, P.M. Nitrofurantoin: Mechanism of action and implications for resistance development in common uropathogens. J. Antimicrob. Chemother. 1994, 33 (Suppl. A), 23–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Wang, F.; Wang, C.; Chen, L.; Liu, H.; Lu, H.; Wen, H.; Zhou, T. Unravelling mechanisms of nitrofurantoin resistance and epidemiological characteristics among Escherichia coli clinical isolates. Int. J. Antimicrob. Agents 2018, 52, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Skerra, A. The Strep-tag system for one-step affinity purification of proteins from mammalian cell culture. Methods Mol. Biol. 2015, 1286, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.G.M.; Skerra, A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007, 2, 1528–1535. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Chiou, J.; Zeng, Z.; Liu, L.; Chen, X.; Zeng, L.; Chan, E.W.C.; Liu, J.-H.; Chen, S. Residues Distal to the Active Site Contribute to Enhanced Catalytic Activity of Variant and Hybrid β-Lactamases Derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 2015, 59, 5976–5983. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Gautier, V.; Saladin-Allard, M.; Hidri, N.; Verdet, C.; Ould-Hocine, Z.; Barnaud, G.; Delisle, F.; Rossier, A.; Lambert, T.; et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 2004, 48, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Naseer, U.; Haldorsen, B.; Tofteland, S.; Hegstad, K.; Scheutz, F.; Simonsen, G.S.; Sundsfjord, A. Norwegian ESBL Study Group Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 2009, 117, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Delmas, J.; Sirot, J.; Shoichet, B.; Bonnet, R. Atomic resolution structures of CTX-M beta-lactamases: Extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 2005, 348, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.J.; Tan, M.X.; Chen, M.I.; Tan, T.Y.; Koo, S.H.; Koong, A.Y.L.; Ng, L.P.; Hu, P.L.; Tan, K.T.; Moey, P.K.S.; et al. Interaction between Antibiotic Resistance, Resistance Genes, and Treatment Response for Urinary Tract Infections in Primary Care. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhai, Y.; Guo, Y.; Li, D.; Wang, Z.; Wang, J.; Chen, Y.; Wang, Q.; Gao, Z. Characterization of Unexpressed Extended-Spectrum Beta-Lactamase Genes in Antibiotic-Sensitive Klebsiella pneumoniae Isolates. Microb. Drug Resist. 2018, 24, 799–806. [Google Scholar] [CrossRef] [PubMed]

| Beta-Lactamase | Hospital 1 | Hospital 2 | Hospital 3 |
|---|---|---|---|
| CTX-M positive samples | 45 | 42 | 44 |
| Number of different CTX-M genes | 12 | 15 | 16 |
| CTX-M-15 frequency | 60% | 64% | 52% |
| CTX-M genes detected | |||
| CTX-M-1 | 1 | 1 | 1 |
| CTX-M-2 | 2 | 1 | 2 |
| CTX-M-3 | 0 | 1 | 1 |
| CTX-M-9 | 1 | 1 | 4 |
| Mutated CTX-M-14 | 5 | 0 | 0 |
| CTX-M-15 | 27 | 27 | 23 |
| CTX-M-16 | 0 | 0 | 1 |
| CTX-M-27 | 1 | 1 | 2 |
| CTX-M-32 | 1 | 1 | 1 |
| CTX-M-51 | 0 | 1 | 0 |
| CTX-M55 | 0 | 1 | 2 |
| CTX-M-59 | 0 | 2 | 1 |
| CTX-M-65 | 0 | 1 | 0 |
| CTX-M-66 | 0 | 1 | 1 |
| CTX-M-90 | 1 | 0 | 0 |
| CTX-M-108 | 1 | 1 | 1 |
| CTX-M-160 | 0 | 0 | 1 |
| CTX-M-163 | 1 | 0 | 1 |
| CTX-M-172 | 1 | 1 | 0 |
| CTX-M-173 | 0 | 0 | 1 |
| CTX-M-203 | 0 | 1 | 0 |
| CTX-M-225 | 0 | 0 | 1 |
| ID | Age | Sex | AMO | CPD | AUG | NIT | TRI | CTX | CAZ | CN | AMI | IMI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 226 | 81 | F | R | R | S | S | R | R | R | S | S | S |
| 246 | 87 | F | R | R | S | R | R | R | R | R | S | S |
| 257 | 91 | F | R | R | S | S | R | R | R | R | S | S |
| 283 | 90 | M | R | R | R | R | R | R | R | R | S | S |
| 294 | 79 | M | R | R | S | S | R | R | S | S | S | S |
| blaCTX-M | Imi | Cfx | Nit | Caz | Ctx | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expression | + | − | + | − | + | − | + | − | + | − |
| CTX-M-15 | 0.23 | 0.004 | 1.8 | 0,047 | 3.6 | 0.064 | 1.16 | 0.023 | 32.0 | 0.073 |
| CTX-M-14 | 0.19 | 0.006 | 1.0 | 0.016 | 6.7 | 0.032 | 1.0 | 0.037 | 1.5 | 0.012 |
| CTX-M-new | 0.064 | 0.002 | 3.0 | 0.016 | 512.0 | 0.037 | 0.83 | 0.016 | 0.75 | 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edowik, Y.; Caspari, T.; Williams, H.M. The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections. Microorganisms 2020, 8, 1983. https://doi.org/10.3390/microorganisms8121983
Edowik Y, Caspari T, Williams HM. The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections. Microorganisms. 2020; 8(12):1983. https://doi.org/10.3390/microorganisms8121983
Chicago/Turabian StyleEdowik, Yasir, Thomas Caspari, and Hugh Merfyn Williams. 2020. "The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections" Microorganisms 8, no. 12: 1983. https://doi.org/10.3390/microorganisms8121983
APA StyleEdowik, Y., Caspari, T., & Williams, H. M. (2020). The Amino Acid Changes T55A, A273P and R277C in the Beta-Lactamase CTX-M-14 Render E. coli Resistant to the Antibiotic Nitrofurantoin, a First-Line Treatment of Urinary Tract Infections. Microorganisms, 8(12), 1983. https://doi.org/10.3390/microorganisms8121983






