Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, Growth Conditions
2.2. Bioreactor Cultivation
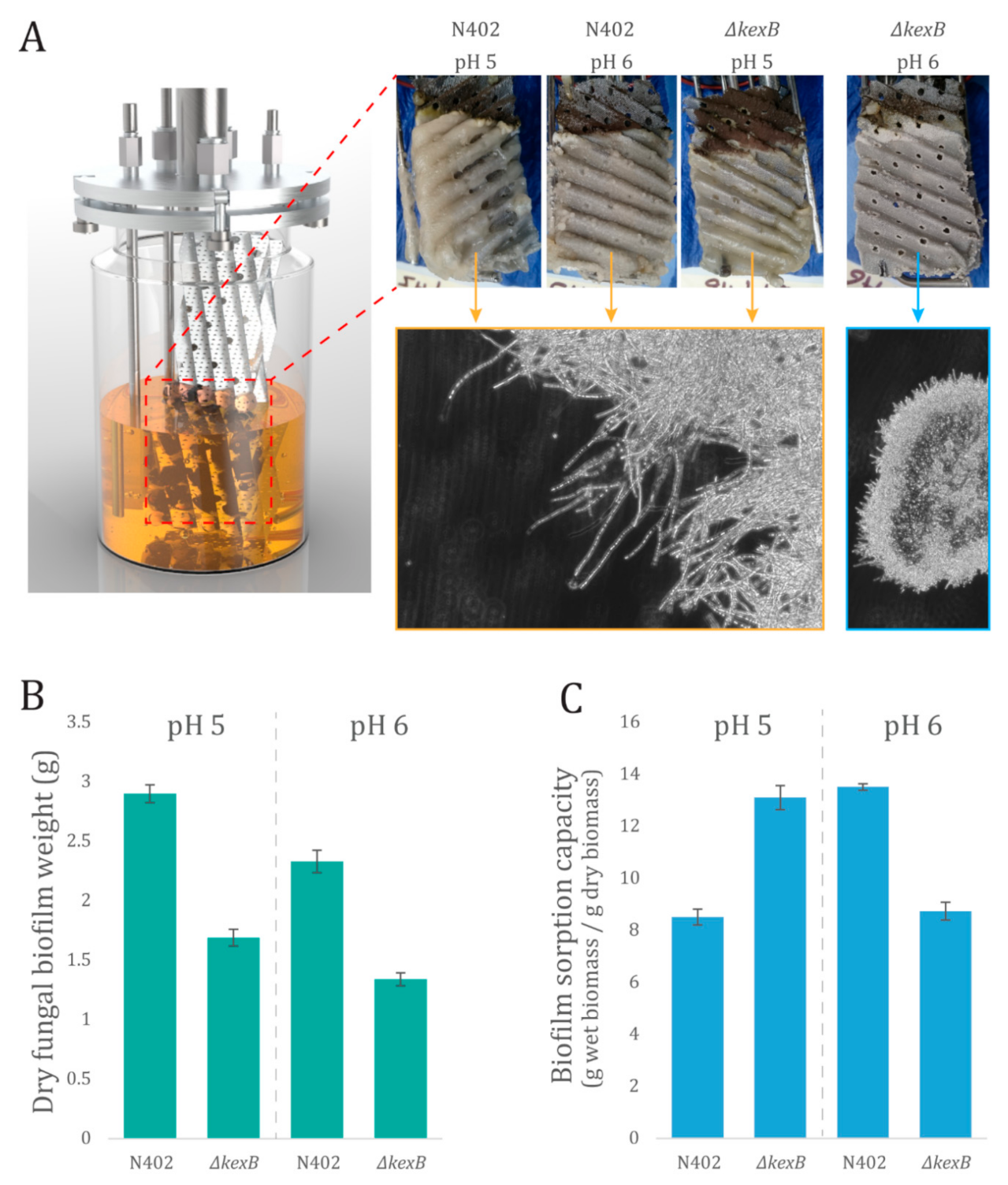
2.3. Biofilm Cultivations
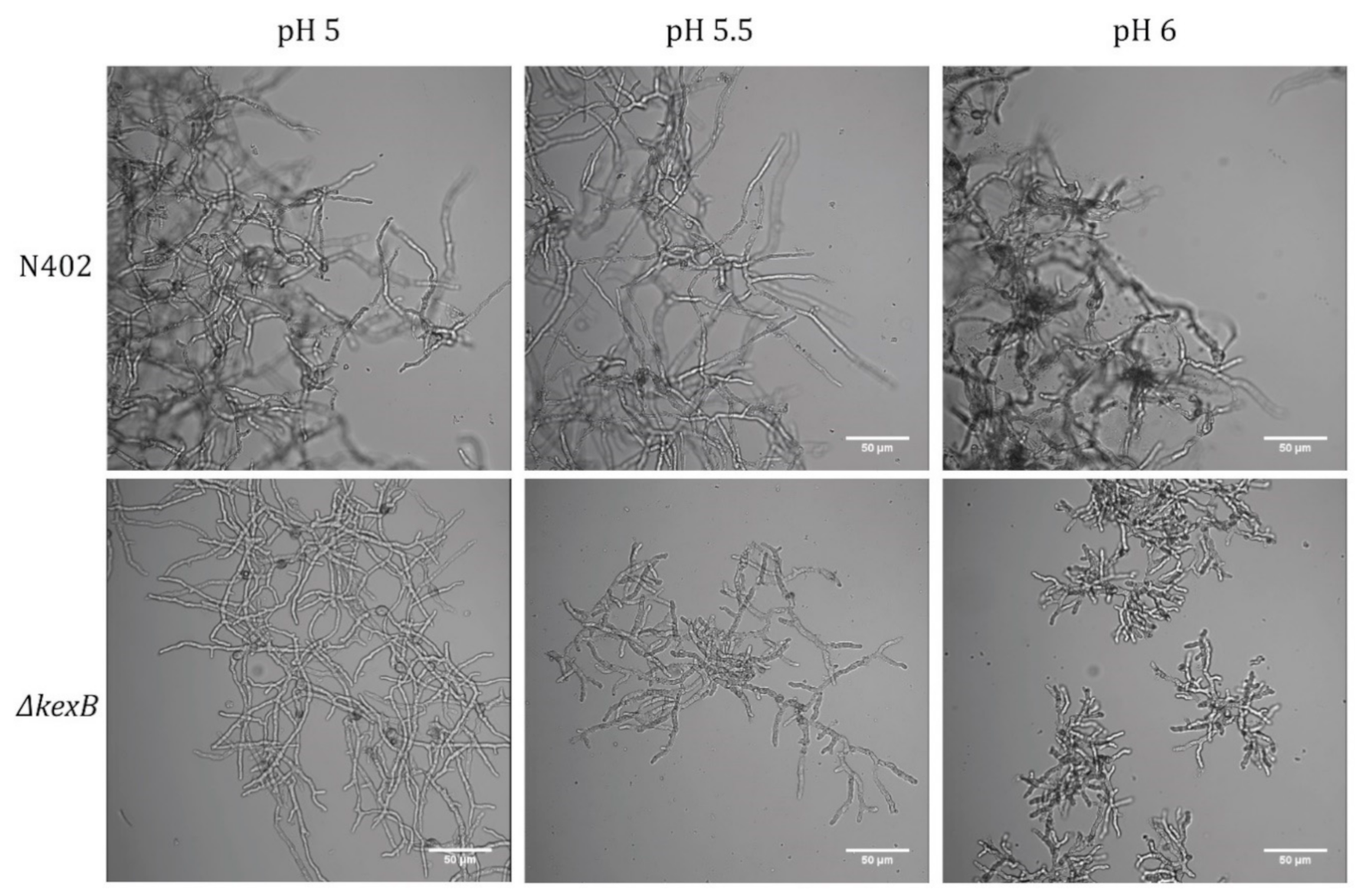
2.4. Microscopy
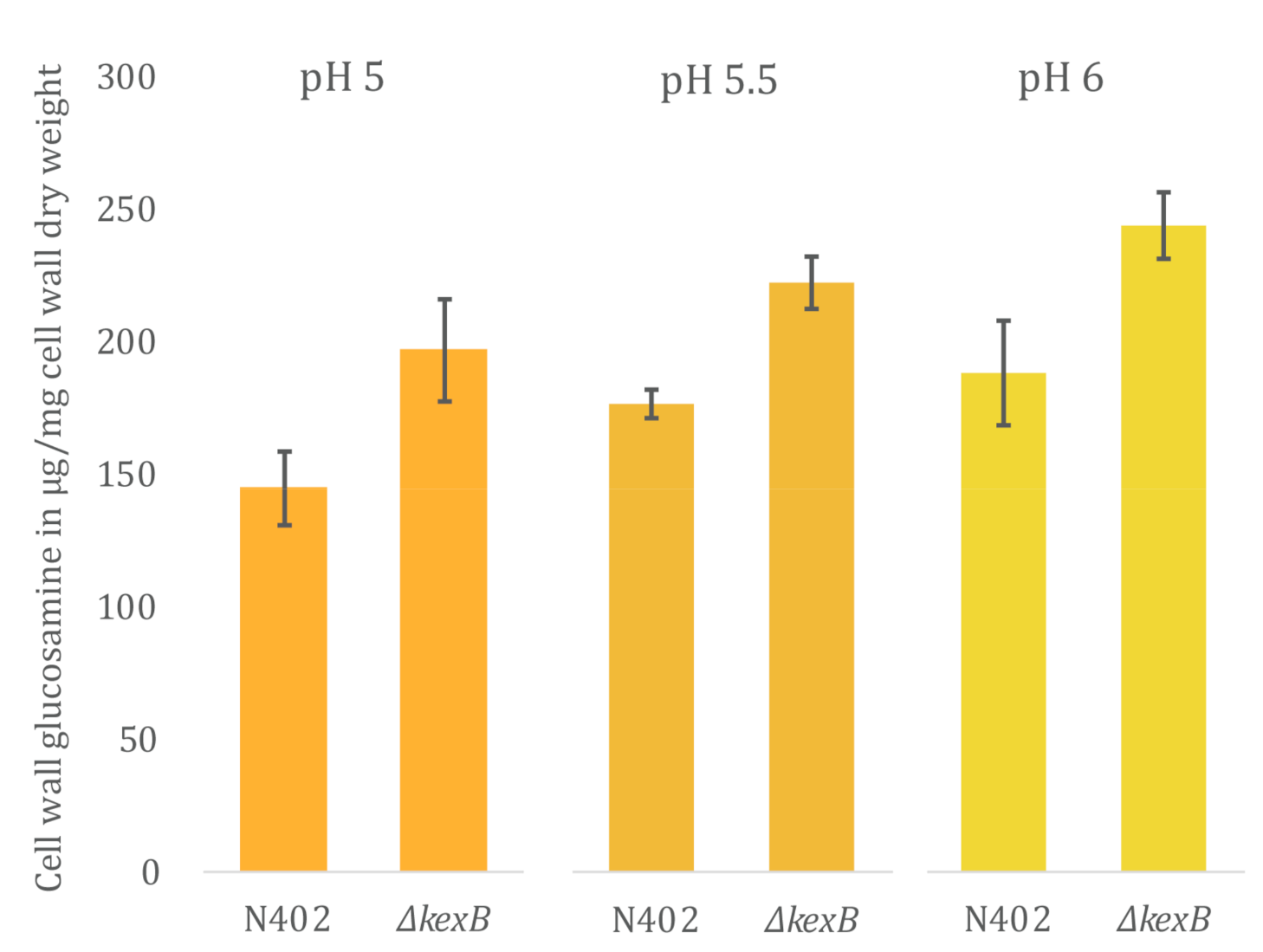
2.5. Cell Wall Isolation and Chitin Analysis
2.6. RNA Isolation and RNA-Sequencing
2.7. Transcriptomic Analysis
2.8. Gene Knockout of kexB in TLF39
2.9. Cell Wall Sensitivity Assays
3. Results
3.1. Disruption of kexB Shows a pH-Dependent Phenotype between pH 5.0 and pH 6.0 in Fermenters
3.2. Disruption of kexB Displays Reduced Biofilm Formation and More Compact Biofilm Structure
3.3. Cell Wall Chitin Content Is Increased in ΔkexB Irrespective of Morphology
3.4. Genome-Wide Expression Profiling Reveals Changes in Expression of Cell Wall Biosynthetic Genes
3.5. Cell Wall Integrity Is Affected by Disruption of kexB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
References
- Orzali, L.; Corsi, B.; Forni, C.; Riccioni, L. Chitosan in Agriculture: A New Challenge for Managing Plant Disease. In Biological Activities and Application of Marine Polysaccharides; InTech: London, UK, 2017. [Google Scholar]
- Naveed, M.; Phil, L.; Sohail, M.; Hasnat, M.; Baig, M.M.F.A.; Ihsan, A.U.; Shumzaid, M.; Kakar, M.U.; Khan, T.M.; Akabar, M.D.; et al. Chitosan oligosaccharide (COS): An overview. Int. J. Biol. Macromol. 2019, 129, 827–843. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, J.; Du, Y.; Fan, L.; Qiu, Y.; Li, J.; Kennedy, J.F. Enzymatic preparation of chitosan from the waste Aspergillus niger mycelium of citric acid production plant. Carbohydr. Polym. 2006, 64, 151–157. [Google Scholar] [CrossRef]
- Nwe, N.; Stevens, W.F. Effect of urea on fungal chitosan production in solid substrate fermentation. Process. Biochem. 2004, 39, 1639–1642. [Google Scholar] [CrossRef]
- Deng, M.-D.; McMullin, T.W.; Grund, A.D. Metabolic engineering for enhanced production of chitin and chitosan in microorganisms. U.S. Patent 2005/0042735A1, 24 February 2005. [Google Scholar]
- Ja’Afaru, M.I. Screening of Fungi Isolated from environmental samples for Xylanase and Cellulase production. ISRN Microbiol. 2013, 283423, 1–7. [Google Scholar] [CrossRef]
- Van Leeuwe, T.M.; Arentshorst, M.; Punt, P.J.; Ram, A.F.J. Interrogation of the cell wall integrity pathway in Aspergillus niger identifies a putative negative regulator of transcription involved in chitin deposition. Gene X 2020, 5, 100028. [Google Scholar] [CrossRef]
- Van Leeuwe, T.M.; Gerritsen, A.; Arentshorst, M.; Punt, P.J.; Ram, A.F.J. Rab GDP-dissociation inhibitor gdiA is an essential gene required for cell wall chitin deposition in Aspergillus niger. Fungal Genet. Biol. 2020, 136, 103319. [Google Scholar] [CrossRef]
- Punt, P.J.; Drint-Kuijvenhoven, A.; Lokman, B.C.; Spencer, J.A.; Jeenes, D.; Archer, D.A.; Van Den Hondel, C.A.M.J.J. The role of the Aspergillus niger furin-type protease gene in processing of fungal proproteins and fusion proteins: Evidence for alternative processing of recombinant (fusion-) proteins. J. Biotechnol. 2003, 106, 23–32. [Google Scholar] [CrossRef]
- Jalving, R.; Van De Vondervoort, P.J.I.; Visser, J.; Schaap, P.J. Characterization of the Kexin-Like Maturase of Aspergillus niger. Appl. Environ. Microbiol. 2000, 66, 363–368. [Google Scholar] [CrossRef][Green Version]
- Fuller, R.S.; Brake, A.; Thorner, J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 1989, 86, 1434–1438. [Google Scholar] [CrossRef]
- Wilcox, C.A.; Fuller, R.S. Posttranslational processing of the prohormone-cleaving Kex2 protease in the Saccharomyces cerevisiae secretory pathway. J. Cell Biol. 1991, 115, 297–307. [Google Scholar] [CrossRef]
- Bryant, N.J.; Stevens, T.H. Two Separate Signals Act Independently to Localize a Yeast Late Golgi Membrane Protein through a Combination of Retrieval and Retention. J. Cell Biol. 1997, 136, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.H.; Wingfield, B.D.; Wingfield, M.; Steenkamp, E. Causes and Consequences of Variability in Peptide Mating Pheromones of Ascomycete Fungi. Mol. Biol. Evol. 2011, 28, 1987–2003. [Google Scholar] [CrossRef] [PubMed]
- Le Marquer, M.; San Clemente, H.; Roux, C.; Savelli, B.; Frei Dit Frey, N. Identification of new signalling peptides through a genome-wide survey of 250 fungal secretomes. BMC Genom. 2019, 20, 64. [Google Scholar]
- Leibowitz, M.J.; Wickner, R.B. A chromosomal gene required for killer plasmid expression, mating, and spore maturation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1976, 73, 2061–2065. [Google Scholar] [CrossRef]
- Wagner, J.C.; Wolf, D.H. Hormone (pheromone) processing enzymes in yeast The carboxy-terminal processing enzyme of the mating pheromone α-factor, carboxypeptidase yscα, is absent in α-factor maturation-defective kex1 mutant cells. FEBS Lett. 1987, 221, 423–426. [Google Scholar] [CrossRef]
- Achstetter, T. Regulation of alpha-factor production in Saccharomyces cerevisiae: A-factor pheromone-induced expression of the MF alpha 1 and STE13 genes. Mol. Cell. Biol. 1989, 9, 4507–4514. [Google Scholar] [CrossRef]
- Heiman, M.G.; Engel, A.; Walter, P. The Golgi-resident protease Kex2 acts in conjunction with Prm1 to facilitate cell fusion during yeast mating. J. Cell Biol. 2007, 176, 209–222. [Google Scholar] [CrossRef]
- Yoshimi, A.; Umemura, M.; Nagano, N.; Koike, H.; Machida, M.; Abe, K. Expression of ustR and the Golgi protease KexB are required for ustiloxin B biosynthesis in Aspergillus oryzae. AMB Express. 2016, 9. [Google Scholar] [CrossRef]
- Te Biesebeke, R.T.; Record, E.; Van Biezen, N.; Heerikhuisen, M.; Franken, A.; Punt, P.J.; van den Hondel, C.A.M.J.J. Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Appl. Microbiol. Biotechnol. 2005, 69, 44–50. [Google Scholar] [CrossRef]
- Newport, G.; Kuo, A.; Flattery, A.; Gill, C.; Blake, J.J.; Kurtz, M.B.; Abruzzo, G.K.; Agabian, N. Inactivation of Kex2p Diminishes the Virulence of Candida albicans. J. Biol. Chem. 2002, 278, 1713–1720. [Google Scholar] [CrossRef]
- Mizutani, O.; Shiina, M.; Yoshimi, A.; Sano, M.; Watanabe, T.; Yamagata, Y.; Nakajima, T.; Gomi, K.; Abe, K. Substantial decrease in cell wall α-1,3-glucan caused by disruption of the kexB gene encoding a subtilisin-like processing protease in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2016, 80, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, O.; Nojima, A.; Yamamoto, M.; Furukawa, K.; Fujioka, T.; Yamagata, Y.; Abe, K.; Nakajima, T. Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae. Eukaryot. Cell 2004, 3, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Jalving, R. Proteolytic Processing in the Secretory Pathway of Aspergillus Niger; Wageningen Universiteit: Wageningen, The Netherlands, 2005. [Google Scholar]
- Arentshorst, M.; Niu, J.; Ram, A.F.J. Efficient generation of Aspergillus niger knock out strains by combining NHEJ mutants and a split marker approach. In Genetic Transformation Systems in Fungi; van den Berg, M.A., Maruthachalam, K., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 1, pp. 263–272. [Google Scholar]
- Bos, C.J.; Debets, A.J.M.; Swart, K.; Huybers, A.; Kobus, G.; Slakhorst, S.M. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 1988, 14, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwe, T.M.; Wattjes, J.; Niehues, A.; Forn-Cuní, G.; Geoffrion, N.; Mélida, H.; Arentshorst, M.; Molina, A.; Tsang, A.; Meijer, A.H.; et al. A seven-membered cell wall related transglycosylase gene family in Aspergillus niger is relevant for cell wall integrity in cell wall mutants with reduced α-glucan or galactomannan. Cell Surf. 2020, 6, 100039. [Google Scholar] [CrossRef]
- Jørgensen, T.R.; Nitsche, B.M.; Lamers, G.E.; Arentshorst, M.; Hondel, C.A.V.D.; Ram, A.F. Transcriptomic insights into the physiology of Aspergillus niger approaching a specific growth rate of zero. Appl. Environ. Microbiol. 2010, 76, 5344–5355. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Aguilar-Pontes, M.; Brandl, J.; McDonnell, E.; Strasser, K.; Nguyen, T.; Riley, R.; Mondo, S.; Salamov, A.; Nybo, J.; Vesth, T.; et al. The gold-standard genome of Aspergillus niger NRRL 3 enables a detailed view of the diversity of sugar catabolism in fungi. Stud. Mycol. 2018, 91, 61–78. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, M.I.L.R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences [version 2; referees: 2 approved]. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Punt, P.J.; Oliver, R.P.; Dingemanse, M.A.; Pouwels, P.H.; Hondel, C.A.V.D. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 1987, 56, 117–124. [Google Scholar] [CrossRef]
- Niu, J.; Arentshorst, M.; Nair, P.D.S.; Dai, Z.; Baker, S.E.; Frisvad, J.C.; Nielsen, K.F.; Punt, P.J.; Ram, A.F.J. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 Genes Genomes Genet. 2016, 6, 193–204. [Google Scholar]
- Zune, Q.; Delepierre, A.; Gofflot, S.; Bauwens, J.; Twizere, J.-C.; Punt, P.J.; Francis, F.; Toye, D.; Boukraa, S.; Delvigne, F. A fungal biofilm reactor based on metal structured packing improves the quality of a Gla:GFP fusion protein produced by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2015, 99, 6241–6254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pel, H.J.; De Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; De Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231. [Google Scholar] [CrossRef]
- Damveld, R.A.; Arentshorst, M.; Franken, A.; vanKuyk, P.A.; Klis, F.M.; van den Hondel, C.A.; Ram, A.F. The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 2005, 58, 305–319. [Google Scholar] [CrossRef]
- Meyer, V.; Damveld, R.A.; Arentshorst, M.; Stahl, U.; Van Den Hondel, C.A.M.J.J.; Ram, A.F.J. Survival in the presence of antifungals: Genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J. Biol. Chem. 2007, 282, 32935–32948. [Google Scholar] [CrossRef]
- Park, J.; Hulsman, M.; Arentshorst, M.; Breeman, M.; Alazi, E.; Lagendijk, E.L.; Rocha, M.C.; Malavazi, I.; Nitsche, B.M.; Hondel, C.A.V.D.; et al. Transcriptomic and molecular genetic analysis of the cell wall salvage response of Aspergillus niger to the absence of galactofuranose synthesis. Cell. Microbiol. 2016, 18, 1268–1284. [Google Scholar] [CrossRef]
- Khalesi, M.; Zune, Q.; Telek, S.; Riveros-Galan, D.; Verachtert, H.; Toye, D.; Gebruers, K.; Derdelinckx, G.; Delvigne, F. Fungal biofilm reactor improves the productivity of hydrophobin HFBII. Biochem. Eng. J. 2014, 88, 171–178. [Google Scholar] [CrossRef]
- Ram, A.F.J.; Arentshorst, M.; Damveld, R.A.; Vankuyk, P.A.; Klis, F.M.; van den Hondel, C.A.M.J.J. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine: Fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 2004, 150, 3315–3326. [Google Scholar] [CrossRef]
- Walker, L.A.; Munro, C.A.; De Bruijn, I.; Lenardon, M.D.; McKinnon, A.D.; Gow, N.A.R. Stimulation of chitin synthesis rescues Candida albicans from Echinocandins. PLoS Pathog. 2008, 4, e1000040. [Google Scholar] [CrossRef]
- Walker, L.A.; Lee, K.K.; Munro, C.A.; Gow, N.A.R.R. Caspofungin treatment of Aspergillus fumigatus results in ChsG-dependent upregulation of chitin synthesis and the formation of chitin-rich microcolonies. Antimicrob. Agents Chemother. 2015, 59, 5932–5941. [Google Scholar] [CrossRef]
- Fortwendel, J.R.; Juvvadi, P.R.; Perfect, B.Z.; Rogg, L.E.; Perfect, J.R.; Steinbach, W.J. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to Caspofungin. Antimicrob. Agents Chemother. 2010, 54, 1555–1563. [Google Scholar] [CrossRef]
- Heilmann, C.J.; Sorgo, A.G.; Mohammadi, S.; Sosinska, G.J.; De Koster, C.G.; Brul, S.; De Koning, L.J.; Klis, F.M. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 2012, 12, 254–264. [Google Scholar] [CrossRef]
- Ram, A.F.J.; Klis, F.M. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 2006, 1, 2253–2256. [Google Scholar] [CrossRef]
- Lagorce, A.; Le Berre-Anton, V.; Aguilar-Uscanga, B.; Martin-Yken, H.; Dagkessamanskaia, A.; François, J. Involvement ofGFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. JBIC J. Biol. Inorg. Chem. 2002, 269, 1697–1707. [Google Scholar] [CrossRef]
- Damveld, R.A.; Franken, A.; Arentshorst, M.; Punt, P.J.; Klis, F.M.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-Galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 2008, 178, 873–881. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Lu, H.; Du, T.; Luo, Y.; Wilson, I.B.H.; Jingyang, W. Kexin-like endoprotease KexB is required for N-glycan processing, morphogenesis and virulence in Aspergillus fumigatus. Fungal Genet. Biol. 2015, 76, 57–69. [Google Scholar] [CrossRef]
- van der Kaaij, R.M.; Yuan, X.L.; Franken, A.; Ram, A.F.; Punt, P.J.; van der Maarel, M.J.; Dijkhuizen, L. Two novel, putatively cell wall-associated and glycosylphosphatidylinositol-anchored alpha-glucanotransferase enzymes of Aspergillus niger. Eukaryot. Cell 2007, 6, 1178–1188. [Google Scholar] [CrossRef]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Function and biosynthesis of cell wall α-1,3-glucan in fungi. J. Fungi 2017, 3, 63. [Google Scholar] [CrossRef]
- Henry, C.; Latgé, J.-P.; Beauvais, A. α1,3 Glucans Are Dispensable in Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 26–29. [Google Scholar] [CrossRef]
- Miyazawa, K.; Yoshimi, A.; Kasahara, S.; Sugahara, A.; Koizumi, A.; Yano, S.; Kimura, S.; Iwata, T.; Sano, M.; Abe, K. Molecular Mass and Localization of α-1,3-Glucan in Cell Wall Control the Degree of Hyphal Aggregation in Liquid Culture of Aspergillus nidulans. Front. Microbiol. 2018, 9, 2623. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Sano, M.; Inaba, A.; Kokubun, Y.; Fujioka, T.; Mizutani, O.; Hagiwara, D.; Fujikawa, T.; Nishimura, M.; Yano, S.; et al. Functional analysis of the α-1,3-Glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB Is the Major α-1,3-Glucan synthase in this fungus. PLoS ONE 2013, 8, e54893. [Google Scholar] [CrossRef]
- Gagnon-Arsenault, I.; Parisé, L.; Tremblay, J.; Bourbonnais, Y. Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 2008, 69, 982–993. [Google Scholar] [CrossRef]
- Miller, K.A.; DiDone, L.; Krysan, D.J. Extracellular secretion of overexpressed glycosylphosphatidylinositol-Linked Cell Wall Protein Utr2/Crh2p as a novel protein quality control mechanism in Saccharomyces cerevisiae. Eukaryot. Cell 2010, 9, 1669–1679. [Google Scholar] [CrossRef][Green Version]
- Grbavac, A.; Čanak, I.; Stuparević, I.; Teparić, R.; Mrša, V. Proteolytic processing of the Saccharomyces cerevisiae cell wall protein Scw4 regulates its activity and influences its covalent binding to glucan. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1864, 507–515. [Google Scholar] [CrossRef]
- Cai, M.; Zhang, Y.; Hu, W.; Shen, W.; Yu, Z.; Zhou, W.; Jiang, T.; Zhou, X.; Zhang, Y. Genetically shaping morphology of the filamentous fungus Aspergillus glaucus for production of antitumor polyketide aspergiolide A. Microb. Cell Factories 2014, 13, 73. [Google Scholar] [CrossRef]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef]
- Dhillon, G.S.; Kaur, S.; Brar, S.K.; Verma, M. Green synthesis approach: Extraction of chitosan from fungus mycelia. Crit. Rev. Biotechnol. 2012, 33, 379–403. [Google Scholar] [CrossRef]

| Name | Genotype | Reference |
|---|---|---|
| N402 | cspA1 | [30]B |
| ∆kexB | cspA1, pyrG-, ΔkexB::AOpyrG (AB4.1∆pclA) | [12] |
| TLF39 | cspA1, ΔkusA::DR-amdS-DR, ΔcrhA-G | [31] |
| TLF69 | cspA1, ΔkusA::DR-amdS-DR, ΔcrhA-G, ΔkexB::hygB | This study |
| Wild Type | ΔkexB | |||
|---|---|---|---|---|
| pH Condition | µmax (h−1) | Max Biomass (gDW·kg−1) | µmax (h−1) | Max Biomass (gDW·kg−1) |
| pH 5.0 | 0.187 | 3.66 | 0.200 | 4.15 |
| pH 5.5 (1) | 0.175 | 3.68 | 0.194 | 4.19 |
| pH 5.5 (2) | 0.167 | 3.54 | 0.180 | 4.30 |
| pH 6.0 | 0.063 * | 0.64 * | 0.211 | 4.31 |
| An ID | NRRL3 ID | Gene Description | Gene | Wild Type (Normalized Read Counts) | ΔkexB (Normalized Read Counts) | FC | Up- or Down-Regulated | Padj |
|---|---|---|---|---|---|---|---|---|
| A-glucan biosynthesis and modification | ||||||||
| An12g02460 | NRRL3_09001 | Putative GPI-anchored amylase-like protein (GH13-family) with possible function in alpha1,3-1,4-glucan processing | agtB | 30 | 1114 | 38.06 | up | 1.43 × 10−69 |
| An12g02450 | NRRL3_09002 | Putative catalytic subunit alpha1,3-glucan synthase complex; SpAgs1-like | agsC | 64 | 1688 | 26.48 | up | 5.91 × 10−67 |
| An04g09890 | NRRL3_07454 | Putative catalytic subunit alpha1,3-glucan synthase complex; SpAgs1-like | agsA * | 104 | 574 | 5.52 | up | 8.90 × 10−25 |
| An08g09610 | NRRL3_11494 | Putative alpha-1,3-glucanase GH71; member of the SpAgn1-family | agnD | 1496 | 6544 | 4.37 | up | 1.55 × 10−71 |
| Β-glucan biosynthesis and modification | ||||||||
| An09g00670 | NRRL3_00054 | Predicted GPI-anchored protein. Putative 1,3-β-glucanosyltransferase GH72; member of the Gel-family | gelD | 40 | 920 | 22.95 | up | 3.83 × 10−73 |
| An03g05290 | NRRL3_08399 | Predicted GPI-anchored protein. Putative beta-1,3-glucanosyltransferase GH17; member of the AfBgt1-family | bgtB | 20,956 | 29,235 | 1.40 | up | 7.26 × 10−4 |
| An01g12450 | NRRL3_02657 | Putative exo-beta-1,3-glucanase (GH55-family); related to Coniothyrium minitans exo-1,3-glucanase (Cmg1) | bxgA * | 4743 | 6407 | 1.35 | up | 8.95 × 10−3 |
| An06g01550 | NRRL3_11624 | Putative catalytic subunit beta1,3-glucan synthase complex; ScFks1-like | fksA | 18,189 | 23,402 | 1.29 | up | 6.29 × 10−4 |
| An07g04650 | NRRL3_04586 | Putative beta-1,3-glucanosyltransferase GH17; member of the AfBgt1-family | bgtC | 1161 | 847 | −1.37 | down | 2.46 × 10−3 |
| An10g00400 | NRRL3_06317 | Predicted GPI-anchored protein. Putative 1,3-β-glucanosyltransferase GH72; A. fumigatus Gel1-like | gelA * | 14,085 | 9570 | −1.47 | down | 4.10 × 10−8 |
| An03g06220 | NRRL3_08332 | Predicted GPI-anchored protein. Putative 1,3-β-glucanosyltransferase GH72; member of the Gel-family | gelE | 2008 | 192 | −10.50 | down | 6.04 × 10−120 |
| An19g00090 | NRRL3_01223 | Putative exo-beta-1,3-glucanase (GH55-family); related to Coniothyrium minitans exo-1,3-glucanase (Cmg1) | bgxC | 1101 | 102 | −10.83 | down | 3.28 × 10−50 |
| Chitin biosynthesis and modification | ||||||||
| N/A | NRRL3_09653 | Putative chitinase (N402 specific) | - | 17 | 336 | 19.52 | up | 2.11 × 10−17 |
| An12g10380 | NRRL3_02932 | Putative chitin synthase ClassIII; EnChsB-like | chsE * | 3764 | 6517 | 1.73 | up | 1.35 × 10−15 |
| An08g05290 | NRRL3_11152 | Putative chitin synthase ClassVI; | chsG | 147 | 252 | 1.71 | up | 4.56 × 10−3 |
| An14g00650 An14g00660 | NRRL3_00641 | Putative chitin synthase ClassI; EnChsC-like | chsC | 2398 | 3814 | 1.59 | up | 2.44 × 10−7 |
| An11g01540 | NRRL3_10021 | Putative transglycosidase of GH16-family involved in cell wall biosynthesis; ScCrh1-like | crhA | 737 | 1097 | 1.49 | up | 8.45 × 10−4 |
| An01g11010 | NRRL3_02532 | Predicted GPI-anchored protein. Putative transglycosidase of GH16-family involved in cell wall biosynthesis; member of the ScCrh1-family | crhD * | 3637 | 5234 | 1.44 | up | 7.65 × 10−4 |
| An09g02290 | NRRL3_00179 | Putative chitin synthase ClassIV; EnChsD-like | chsD | 2278 | 3104 | 1.36 | up | 1.99 × 10−3 |
| An09g04010 | NRRL3_00331 | Putative chitin synthase ClassIII; EnChsB-like | chsB * | 6774 | 8553 | 1.26 | up | 5.98 × 10−3 |
| An01g05360 | NRRL3_02063 | Putative ClassV Chitinase (GH18); ScCts2-like | cfcD | 1384 | 818 | −1.69 | down | 3.96 × 10−5 |
| An16g02850 | NRRL3_07085 | Putative transglycosidase of GH16-family involved in cell wall biosynthesis; member of the ScCrh1-family | crhF | 1468 | 812 | −1.81 | down | 4.92 × 10−8 |
| An07g07530 | NRRL3_04809 | Predicted GPI-anchored protein. Putative transglycosidase of GH16-family involved in cell wall biosynthesis; ScCrh2-like | crhB | 2205 | 988 | −2.23 | down | 1.53 × 10−9 |
| N/A | NRRL3_04221 | Putative chitinase (N402 specific) | - | 174 | 58 | −2.99 | 3.09 × 10−3 | |
| An19g00100 | NRRL3_01224 | Putative ClassV Chitinase (GH18); ScCts2-like | cfcG | 415 | 2 | −197.88 | 8.05 × 10−20 | |
| GH76 family proteins | ||||||||
| An11g01240 | NRRL3_10041 | Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation; ScDfg5-like | dfgH | 289 | 1103 | 3.82 | up | 2.58 × 10−35 |
| An14g03520 | NRRL3_00897 | Predicted GPI-anchored protein. Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation; ScDfg5-like | dfgC * | 1133 | 1996 | 1.76 | up | 6.95 × 10−10 |
| An16g08090 | NRRL3_06700 | Predicted GPI-anchored protein. Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation; ScDfg5-like | dfgE | 886 | 1169 | 1.32 | up | 2.14 × 10−2 |
| An02g02660 | NRRL3_06048 | Putative endo-mannanase (GH76-family) with a possible role in GPI-CWP incorporation; ScDfg5-like | dfgG | 2587 | 1693 | −1.53 | down | 3.75 × 10−5 |
| Rho-GAPs | ||||||||
| An18g06730 | NRRL3_10703 | Putative Cdc42-GTPase Activating protein (GAP) with similarity to ScBem3p | capB | 1379 | 1783 | 1.29 | up | 5.54 × 10−3 |
| An13g00850 | NRRL3_01500 | Putative Rho1-GTPase Activating protein (GAP) with strong similarity to ScRgd2 | rapE | 2256 | 2755 | 1.22 | up | 1.91 × 10−2 |
| CWI signaling | ||||||||
| An04g10140 | NRRL3_07436 | Putative plasma membrane sensor required for cell wall integrity signaling; ScMtl1like | mtlB | 93 | 575 | 6.16 | up | 6.51 × 10−22 |
| An07g04070 | NRRL3_04545 | Putative plasma membrane sensor-transducer of the stress-activated PKC1-MPK1 kinase pathway involved in maintenance of cell wall integrity; ScWsc1-like | wscB * | 2073 | 2963 | 1.43 | up | 4.60 × 10−5 |
| An18g02400 | NRRL3_10351 | Protein kinase C with putative function in CWI signaling | pkcA | 3602 | 4518 | 1.25 | up | 5.43 × 10−3 |
| An08g10670 | NRRL3_11584 | MAPK with putative function in Pheromone response/pseudohyphal growth pathway; ScFus3-like | fusC | 2269 | 2808 | 1.24 | up | 1.39 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Leeuwe, T.M.; Arentshorst, M.; Forn-Cuní, G.; Geoffrion, N.; Tsang, A.; Delvigne, F.; Meijer, A.H.; Ram, A.F.J.; Punt, P.J. Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content. Microorganisms 2020, 8, 1918. https://doi.org/10.3390/microorganisms8121918
van Leeuwe TM, Arentshorst M, Forn-Cuní G, Geoffrion N, Tsang A, Delvigne F, Meijer AH, Ram AFJ, Punt PJ. Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content. Microorganisms. 2020; 8(12):1918. https://doi.org/10.3390/microorganisms8121918
Chicago/Turabian Stylevan Leeuwe, Tim M., Mark Arentshorst, Gabriel Forn-Cuní, Nicholas Geoffrion, Adrian Tsang, Frank Delvigne, Annemarie H. Meijer, Arthur F. J. Ram, and Peter J. Punt. 2020. "Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content" Microorganisms 8, no. 12: 1918. https://doi.org/10.3390/microorganisms8121918
APA Stylevan Leeuwe, T. M., Arentshorst, M., Forn-Cuní, G., Geoffrion, N., Tsang, A., Delvigne, F., Meijer, A. H., Ram, A. F. J., & Punt, P. J. (2020). Deletion of the Aspergillus niger Pro-Protein Processing Protease Gene kexB Results in a pH-Dependent Morphological Transition during Submerged Cultivations and Increases Cell Wall Chitin Content. Microorganisms, 8(12), 1918. https://doi.org/10.3390/microorganisms8121918








