Identification of an Antiviral Compound from the Pandemic Response Box that Efficiently Inhibits SARS-CoV-2 Infection In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Compound Preparation of the Pandemic Response Box
2.3. Antiviral Activity Screening
2.4. Cell Cytotoxicity and Cell Viability
2.5. Half Maximal Effective Concentration (EC50) Determination of Selected Compounds
2.6. Half-Maximum Cytotoxicity Concentration (CC50) Determination
2.7. Time-of-Addition Experiment
2.8. Data Representation
3. Results
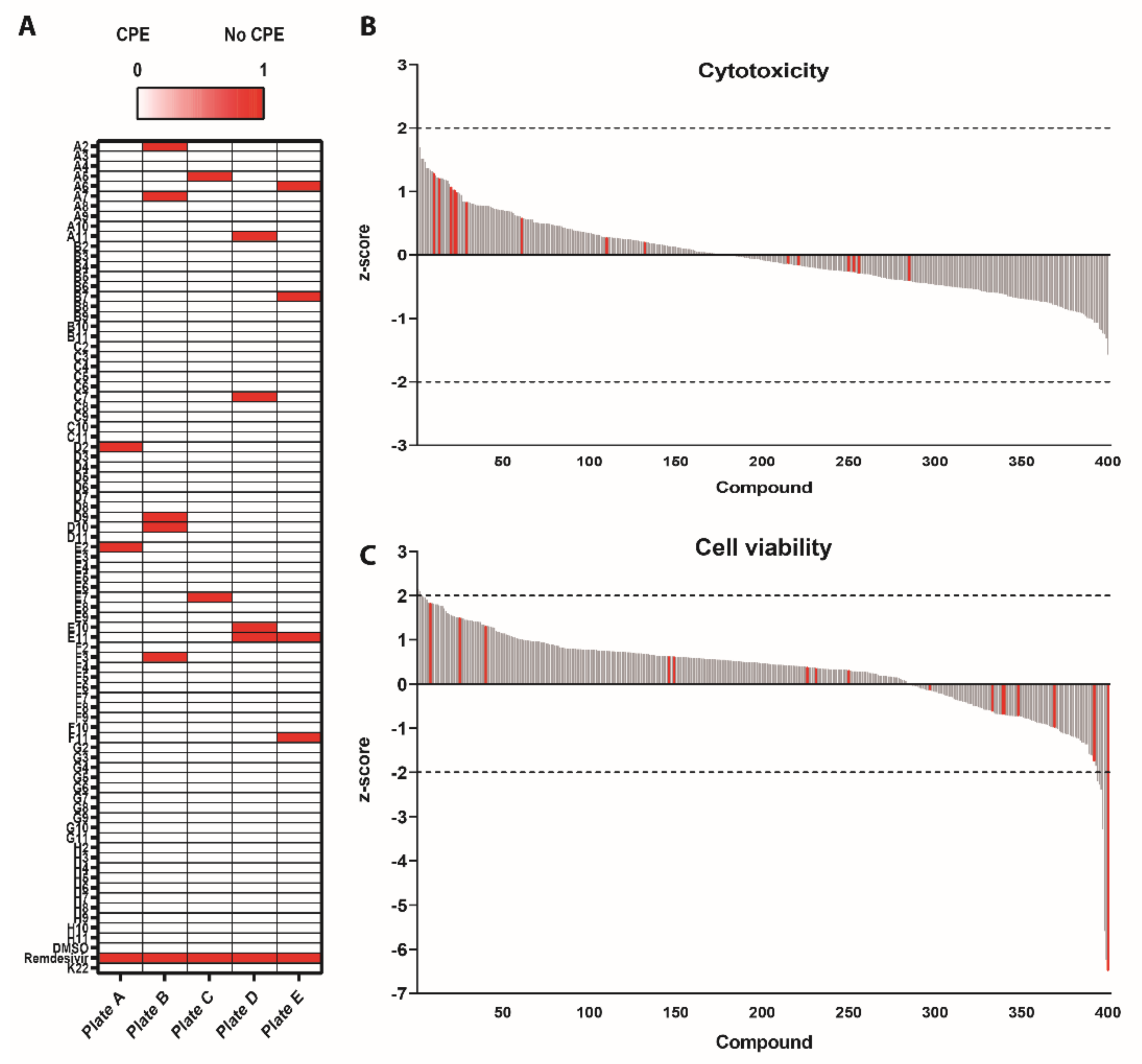
3.1. Survival Screen with Compounds Included in the Pandemic Response Box against SARS-CoV-2
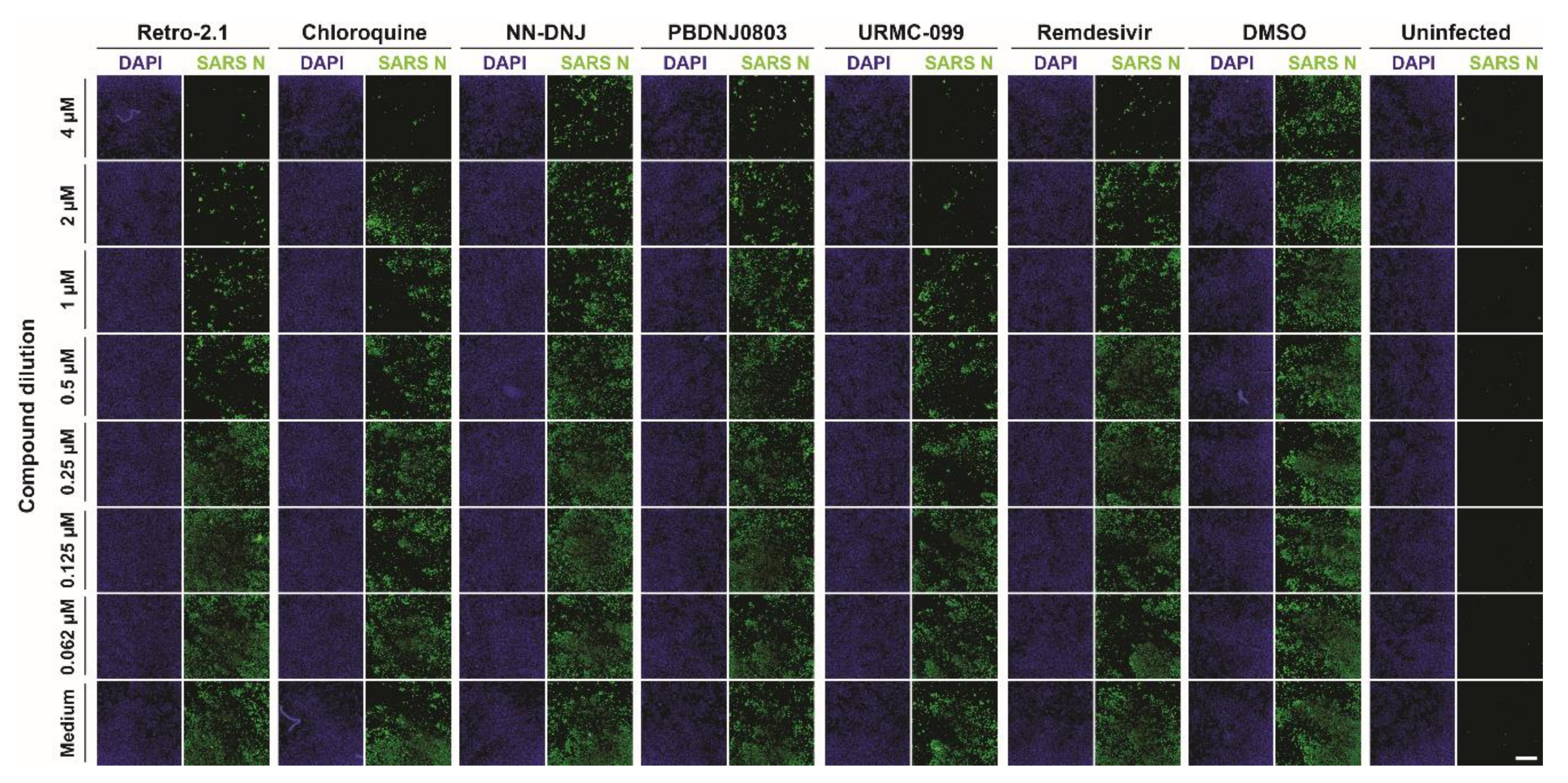
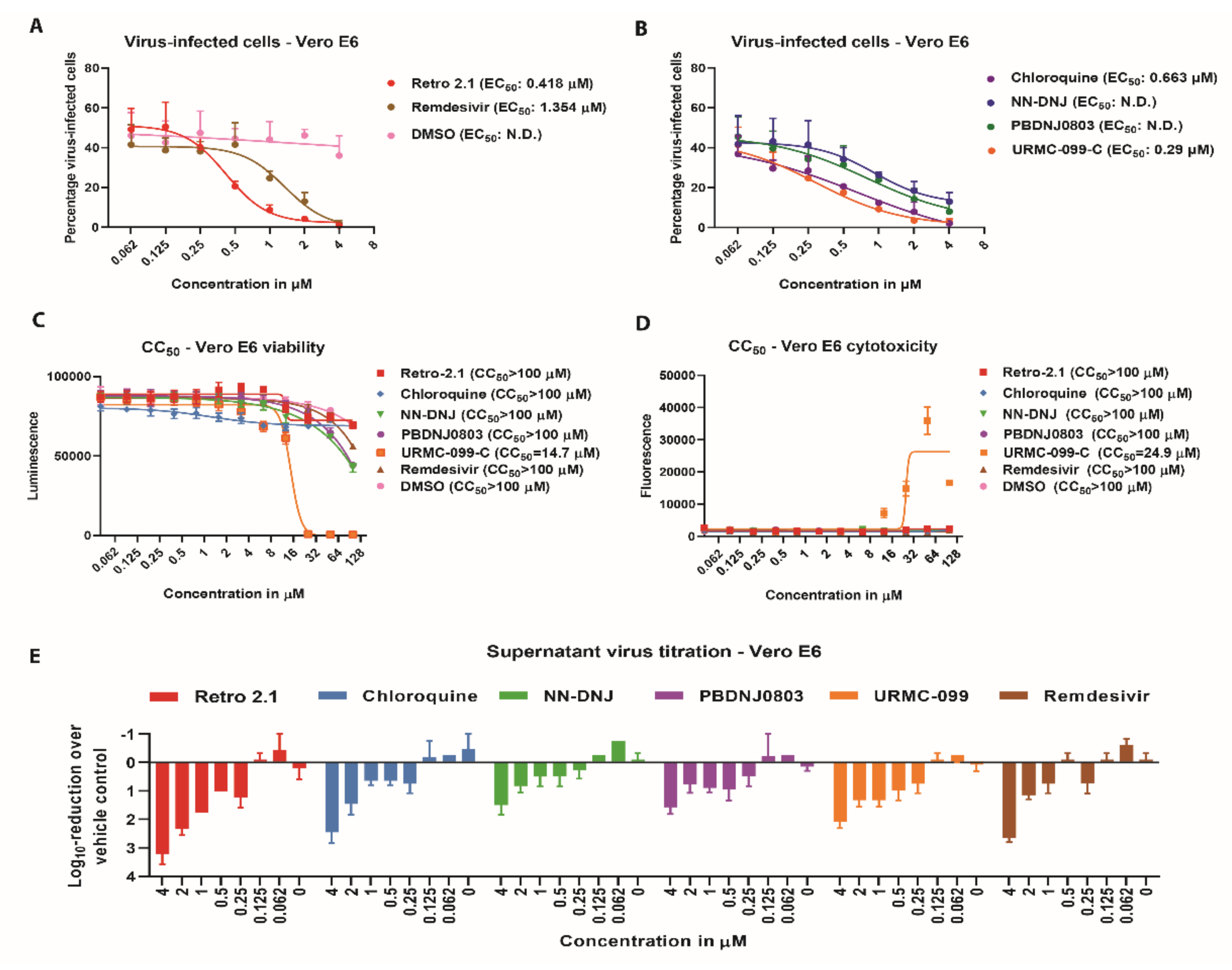
3.2. Antiviral Efficacy against SARS-CoV-2
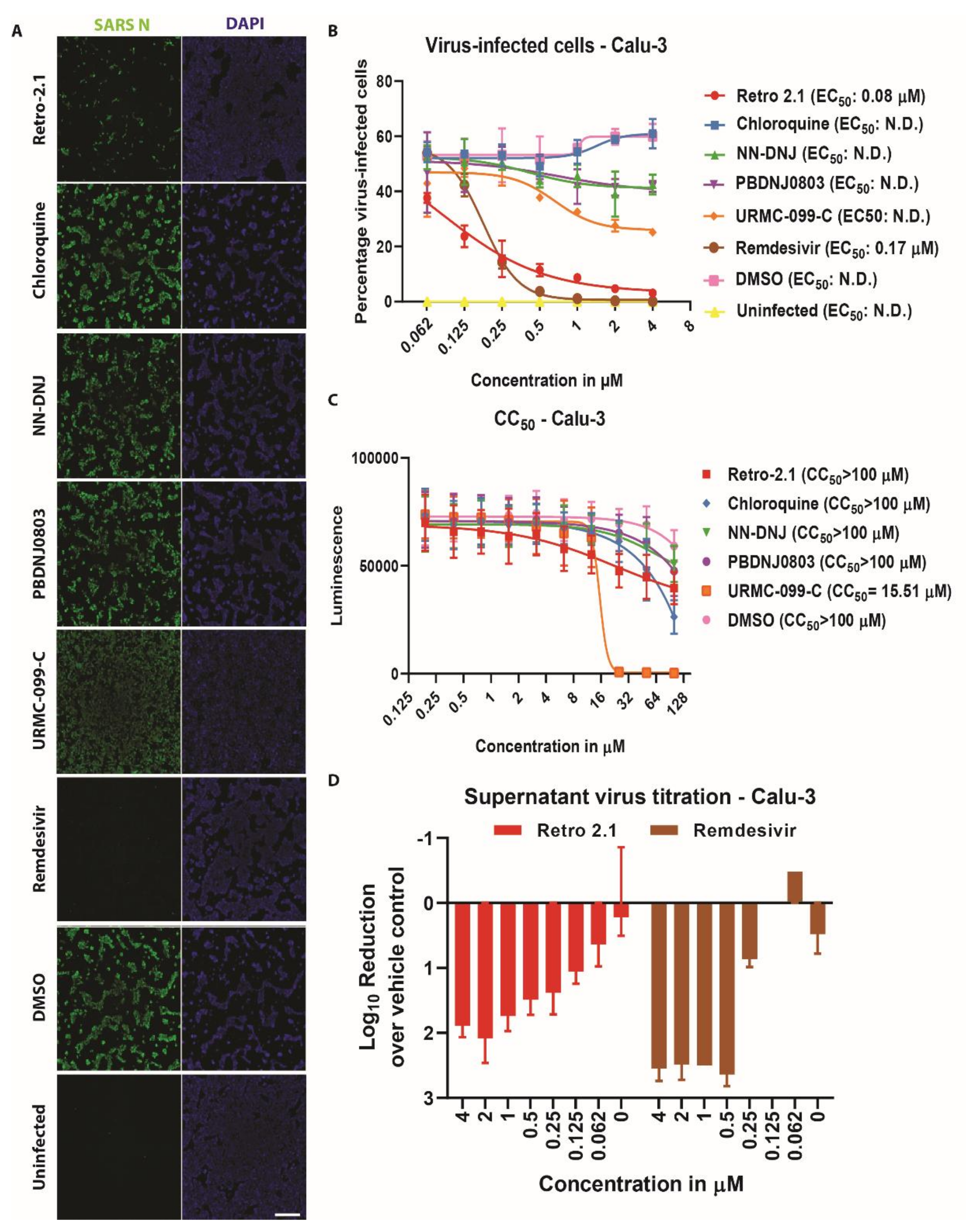
3.3. Inhibition Analysis of SARS-CoV-2 Infection on Calu-3 Cells
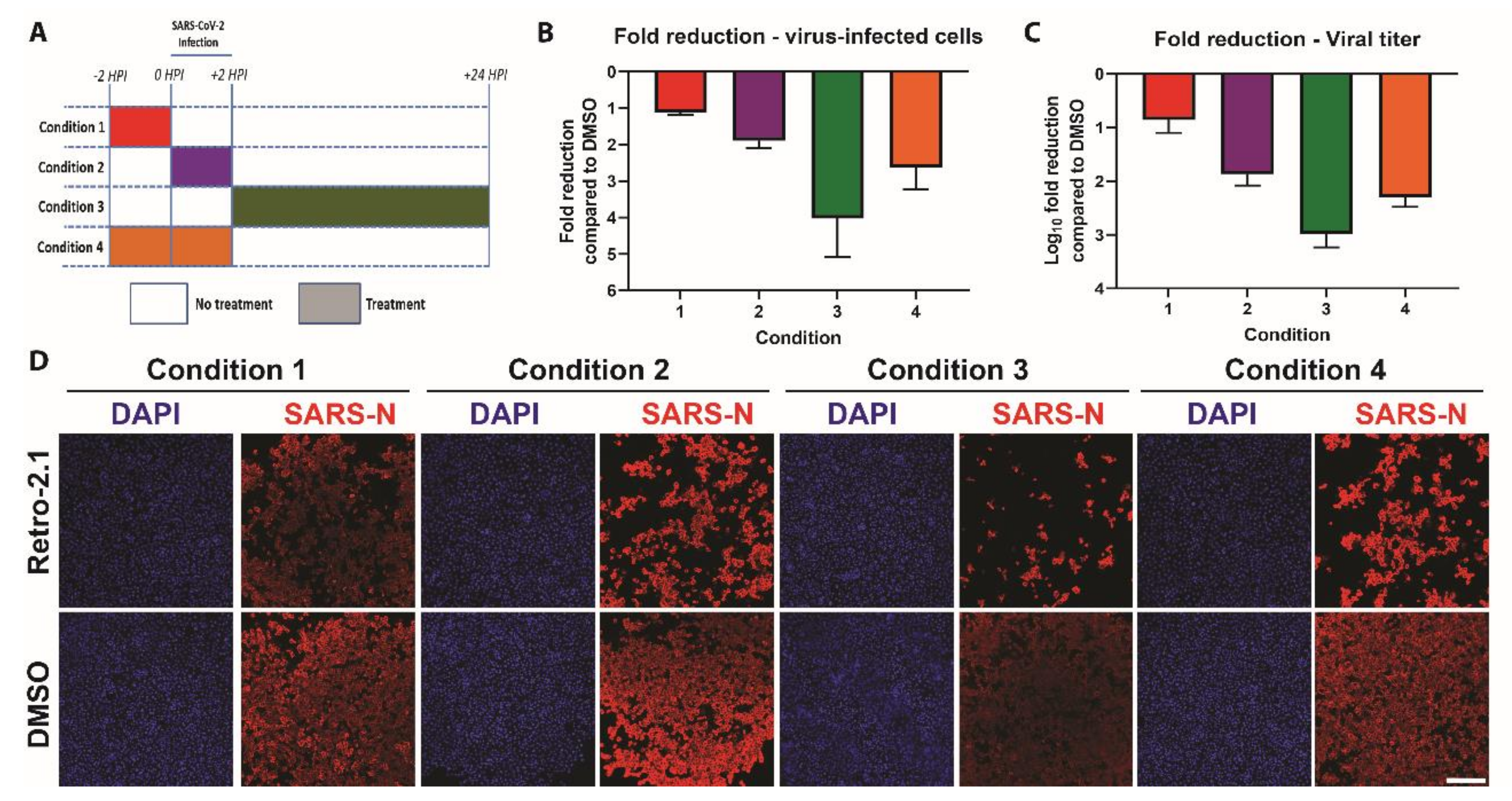
3.4. Time-of-Addition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Plate Position: | Section | MVV-Number | CHEMBL ID | Reference | Name | Clinical Stage According CHEMBL | Chemical Formula |
|---|---|---|---|---|---|---|---|
| Plate A, D2 | Antifungals | MMV1634386 | CHEMBL3311228 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL3311228/ | Oteseconazole | 3 (Phase III) | C23 H16 F7 N5 O2 |
| Plate A, E2 | Antifungals | MMV637528 | CHEMBL64391 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL64391/ | Itraconazole | 4 (approved) | C35 H38 Cl2 N8 O4 |
| Plate B, A2 | Antibacterials | MMV1483032 | CHEMBL243644 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL243644/ | AC1MTT7T | 0 (research) | C22 H18 N4 O4 |
| Plate B, A7 | Antibacterials | MMV637945 | CHEMBL403 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL403/ | Sulbactam | 4 (approved) | C8 H11 N O5 S |
| Plate B, D9 | Antibacterials | MMV1582492 | CHEMBL3109593 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL3109593/ | Retro-2.1 | 0 (research) | C23 H18 F N3 O S2 |
| Plate B, F3 | Antibacterials | MMV1582487 | CHEMBL198796 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL198796/ | Decylphosphinate | 0 (research) | C13 H28 N O4 P |
| Plate C, A5 | Antibacterials | MMV1578576 | CHEMBL1568820 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL1568820/ | - | 0 (research) | C15 H12 F N3 O |
| Plate C, E7 | Antibacterials | MMV000008 | CHEMBL76 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL76/ | Chloroquine | 4 (approved) | C18 H26 Cl N3 |
| Plate D, A11 | Antivirals | MMV1593513 | CHEMBL408500 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL408500/ | N-Nonyl Deoxynojirimycin (NN-DNJ) | 0 (research) | C15 H31 N O4 |
| Plate D, C7 | Antivirals | MMV690621 | - | Patent: WO2006118607A2 | NA for racemic | - | C18 H16 Cl N3 O |
| Plate D, E10 | Antivirals | MMV1634401 | CHEMBL1652119 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL1652119/ | PBDNJ0803 | 0 (research) | C20 H33 N O5 |
| Plate D, E11 | Antivirals | MMV002780 | CHEMBL402487 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL402487/ | Noscapine | 0 (research) | C22 H23 N O7 |
| Plate E, A6 | Antivirals | MMV1580482 | CHEMBL2436978 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL2436978/ Patent: WO 2014085795 A1 | URMC-099-C | 0 (research) | C27 H27 N5 |
| Plate E, B7 | Antivirals | MMV1593544 | CHEMBL3752642 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL3752642/ | - | 0 (research) | C36 H43 N3 O5 |
| Plate E, E11 | Antifungals | MMV002350 | CHEMBL561 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL561/ | Lomefloxacin | 4 (approved) | C17H19F2N3O3 |
| Plate E, F11 | Antivirals | MMV1580483 | CHEMBL3960662 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL3960662/ | AZD0156 | 0 (research) | C26 H31 N5 O3 |
| - | Control | - | CHEMBL4065616 | https://www.ebi.ac.uk/chembl/compound_report_card/CHEMBL4065616/ | Remdesivir | 2 (Phase II) | C27 H35 N6 O8 P |
Appendix B
| Compound | EC50 in μM | EC90 in μM | CC50 in μM | Selective Index (SI) | Class | Examples of Susceptible Viruses | Literature Reference | Mode of Action | Administration | Tested on Cells or Organism | Molecular Structure |
|---|---|---|---|---|---|---|---|---|---|---|---|
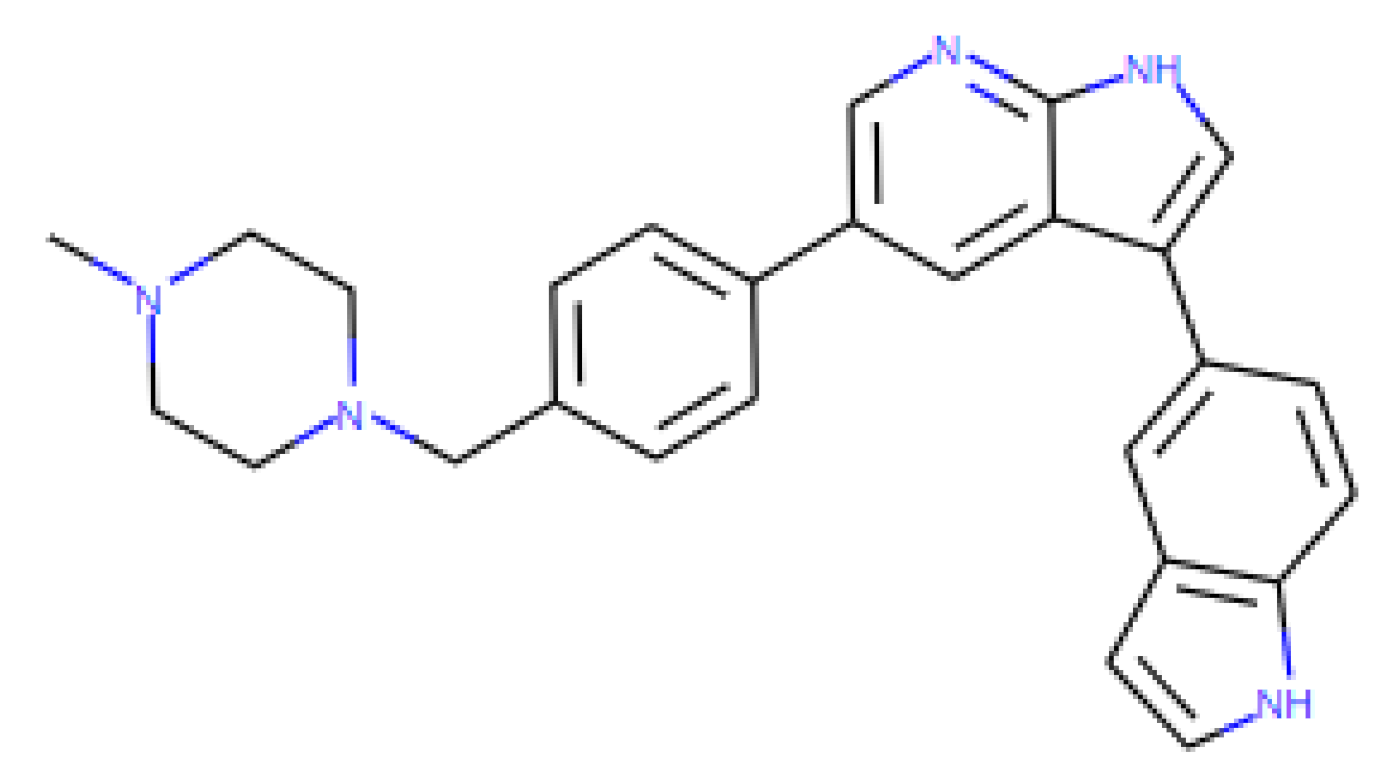
| Retro-2.1 | 0.418 | 1.03 | 100 | 239.2 | Retrograde transport inhibitor |
| [31,32,33,34,35,36] |
|
|
|  |
| Chloroquine | 0.663 | 10.44 | 100 | 150.82 | Autophagic proteolysis inhibitor Endosomal acidification inhibitor |
| [40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62] |
|
|
|  |
| N-Nonyl Deoxyno Jirimycin (NNDNJ) | N.D. | N.D. | 100 | N.D. | ER α-glucosidase inhibitors |
| [63,64,65,66,67,68,69,70,71] US patent: 9040488 |
|
|
| 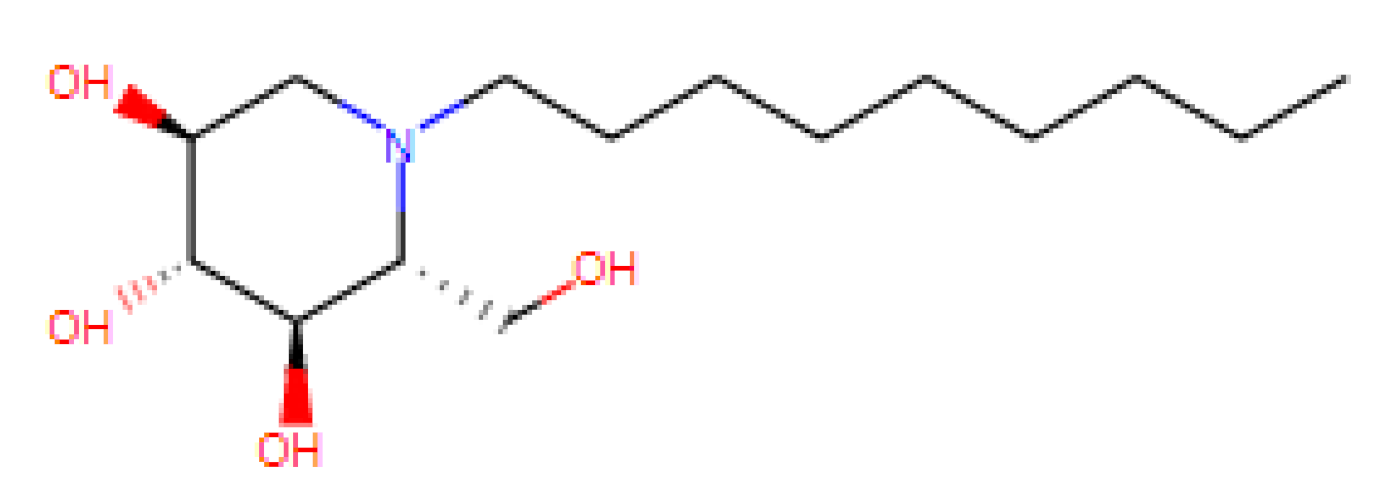 |
| PBDNJ0803 | N.D. | N.D. | 100 | N.D. | ER α-glucosidase inhibitors |
| [69,70] US Patent: 9040488 |
|
|
| 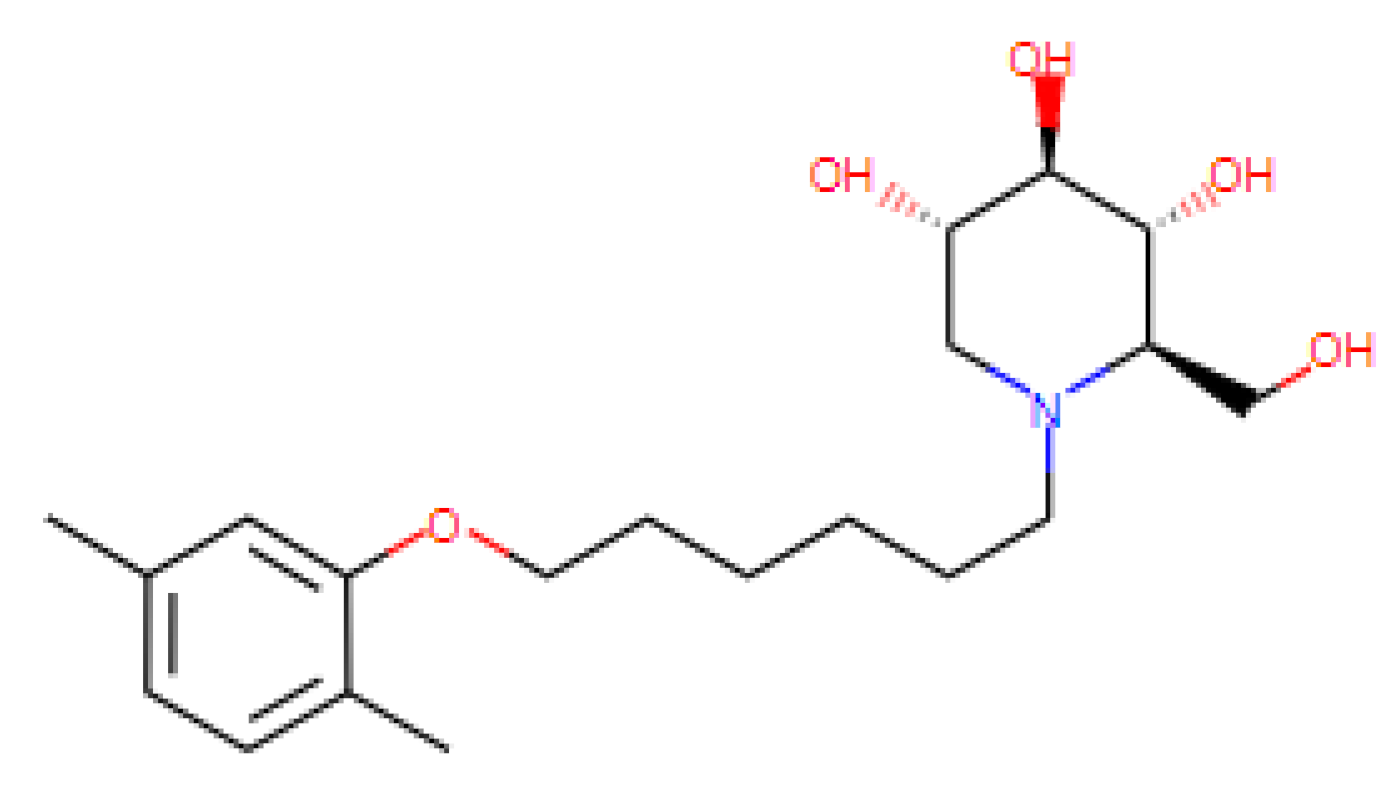 |
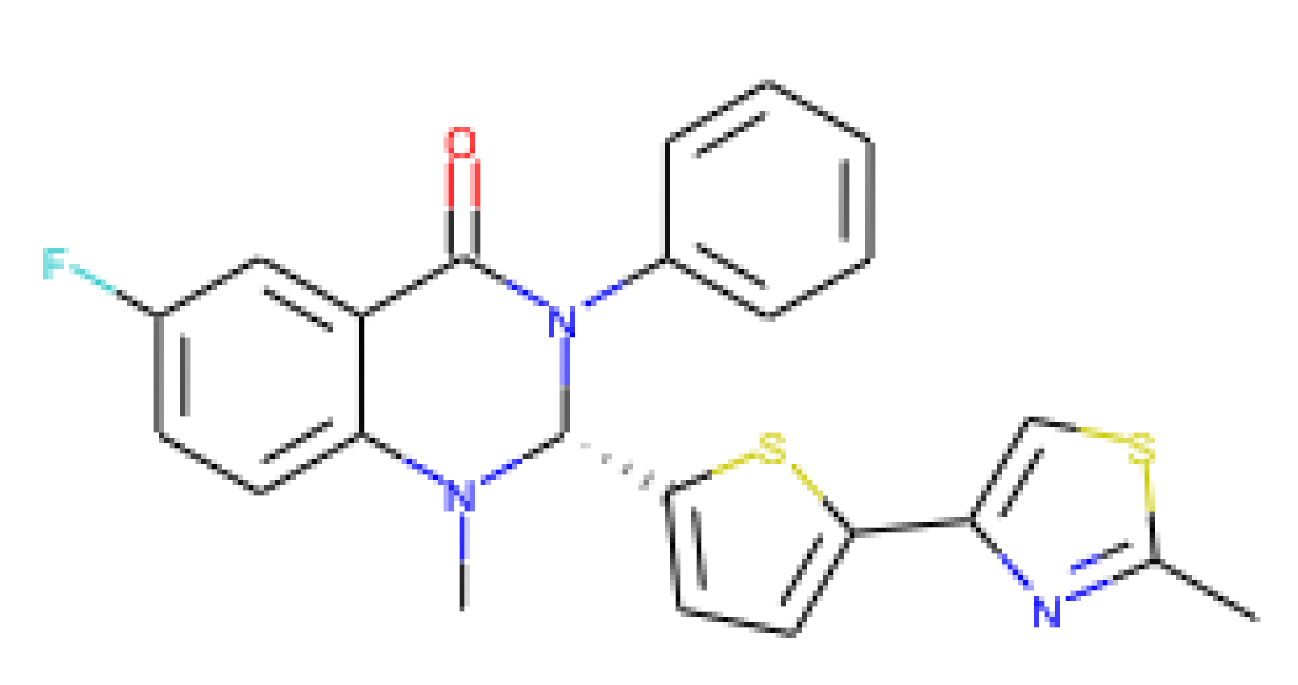
| URMC-099-C | 0.29 | 1.83 | 14.7 | 50.69 | Mixed-lineage kinase 3 inhibitor |
| [72,73,74,75] |
|
|
|  |
| Remdesivir | 1.354 | 3.39 | 100 | 73.85 | Nucleoside analogue |
| [18,76,77,78,79,80] |
|
|
|  |
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 94, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Disease (COVID-19) Situation Reports. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 18 November 2020).
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020, 382, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Home—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 28 October 2020).
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Final Report. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. N. Engl. J. Med. 2020, 383, 517–525. [Google Scholar] [CrossRef]
- Geleris, J.; Sun, Y.; Platt, J.; Zucker, J.; Baldwin, M.; Hripcsak, G.; Labella, A.; Manson, D.K.; Kubin, C.; Barr, R.G.; et al. Observational study of hydroxychloroquine in hospitalized patients with COVID-19. N. Engl. J. Med. 2020, 382, 2411–2418. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
- Pan, H.; Peto, R.; Karim, Q.A.; Alejandria, M.; Henao-Restrepo, A.M.; García, C.H.; Kieny, M.-P.; Malekzadeh, R.; Murthy, S.; Preziosi, M.-P.; et al. Repurposed antiviral drugs for COVID-19 –interim WHO SOLIDARITY trial results. MedRxiv 2020. [Google Scholar] [CrossRef]
- Smith, E.C.; Blanc, H.; Vignuzzi, M.; Denison, M.R. Coronaviruses Lacking Exoribonuclease Activity Are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PLoS Pathog. 2013, 9, e1003565. [Google Scholar] [CrossRef]
- Lundin, A.; Dijkman, R.; Bergström, T.; Kann, N.; Adamiak, B.; Hannoun, C.; Kindler, E.; Jónsdóttir, H.R.; Muth, D.; Kint, J.; et al. Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus. PLoS Pathog. 2014, 10, e1004166. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Bloom, D.E.; Cadarette, D. Infectious Disease Threats in the Twenty-First Century: Strengthening the Global Response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef]
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 18 November 2020).
- WHO | World Now at the Start of 2009 Influenza Pandemic. Available online: https://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/ (accessed on 18 November 2020).
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; V’kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratze, A.; et al. 9 Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020. [Google Scholar] [CrossRef]
- Mutterer, J.; Zinck, E. Quick-and-clean article figures with FigureJ. J. Microsc. 2013, 252, 89–91. [Google Scholar] [CrossRef]
- Rappe, J.C.F.; de Wilde, A.; Di, H.; Müller, C.; Stalder, H.; V’kovski, P.; Snijder, E.; Brinton, M.A.; Ziebuhr, J.; Ruggli, N.; et al. Antiviral activity of K22 against members of the order Nidovirales. Virus Res. 2018, 246, 28–34. [Google Scholar] [CrossRef]
- García-Nicolás, O.; V’kovski, P.; Vielle, N.J.; Ebert, N.; Züst, R.; Portmann, J.; Stalder, H.; Gaschen, V.; Vieyres, G.; Stoffel, M.; et al. The small-compound inhibitor K22 displays broad antiviral activity against different members of the family Flaviviridae and offers potential as a panviral inhibitor. Antimicrob. Agents Chemother. 2018, 62, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Mösbauer, K.; Hofmann-Winkler, H.; Kaul, A.; Kleine-Weber, H.; Krüger, N.; Gassen, N.C.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 2020, 585, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Pruijssers, A.J.; George, A.S.; Schäfer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H.; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32, 107940. [Google Scholar] [CrossRef] [PubMed]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; van Helden, J.; Decroly, E.; de Lamballerie, X.; Coutard, B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef]
- Rosenke, K.; Jarvis, M.; Feldmann, F.; Schwarz, B.; Okumura, A.; Lovaglio, J.; Saturday, G.; Hanley, P.; Meade-White, K.; Williamson, B.; et al. Hydroxychloroquine Proves Ineffective in Hamsters and Macaques Infected with SARS-CoV-2. BioRxiv Prepr. Serv. Biol. 2020. [Google Scholar] [CrossRef]
- Dai, W.; Wu, Y.; Bi, J.; Lu, X.; Hou, A.; Zhou, Y.; Sun, B.; Kong, W.; Barbier, J.; Cintrat, J.C.; et al. Antiviral effects of Retro-2cycl and Retro-2.1 against Enterovirus 71 in vitro and in vivo. Antivir. Res. 2017, 144, 311–321. [Google Scholar] [CrossRef]
- Shtanko, O.; Sakurai, Y.; Reyes, A.N.; Noël, R.; Cintrat, J.C.; Gillet, D.; Barbier, J.; Davey, R.A. Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection. Antivir. Res. 2018, 149, 154–163. [Google Scholar] [CrossRef]
- Dai, W.; Wu, Y.; Bi, J.; Wang, J.; Wang, S.; Kong, W.; Barbier, J.; Cintrat, J.C.; Gao, F.; Jiang, Z.; et al. Antiviral effect of Retro-2.1 against herpes simplex Virus type 2 in Vitro. J. Microbiol. Biotechnol. 2018, 28, 849–859. [Google Scholar] [CrossRef]
- Harrison, K.; Haga, I.R.; Pechenick Jowers, T.; Jasim, S.; Cintrat, J.-C.; Gillet, D.; Schmitt-John, T.; Digard, P.; Beard, P.M. Vaccinia Virus Uses Retromer-Independent Cellular Retrograde Transport Pathways To Facilitate the Wrapping of Intracellular Mature Virions during Virus Morphogenesis. J. Virol. 2016, 90, 10120–10132. [Google Scholar] [CrossRef]
- Maru, S.; Jin, G.; Desai, D.; Amin, S.; Shwetank; Lauver, M.D.; Lukacher, A.E. Inhibition of Retrograde Transport Limits Polyomavirus Infection In Vivo. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Desai, D.; Lauver, M.; Ostman, A.; Cruz, L.; Ferguson, K.; Jin, G.; Roper, B.; Brosius, D.; Lukacher, A.; Amin, S.; et al. Inhibition of diverse opportunistic viruses by structurally optimized retrograde trafficking inhibitors. Bioorganic Med. Chem. 2019, 27, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Pons, V.; Noël, R.; Buisson, D.A.; Michau, A.; Johannes, L.; Gillet, D.; Barbier, J.; Cintrat, J.C. (S)-N-methyldihydroquinazolinones are the active enantiomers of retro-2 derived compounds against toxins. ACS Med. Chem. Lett. 2014, 5, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Stechmann, B.; Bai, S.K.; Gobbo, E.; Lopez, R.; Merer, G.; Pinchard, S.; Panigai, L.; Tenza, D.; Raposo, G.; Beaumelle, B.; et al. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 2010, 141, 231–242. [Google Scholar] [CrossRef] [PubMed]
- V’kovski, P.; Gerber, M.; Kelly, J.; Pfaender, S.; Ebert, N.; Braga Lagache, S.; Simillion, C.; Portmann, J.; Stalder, H.; Gaschen, V.; et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife 2019, 8, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Wu, Y.; Bi, J.; Wang, S.; Li, F.; Kong, W.; Barbier, J.; Cintrat, J.C.; Gao, F.; Gillet, D.; et al. Antiviral effects of abma against herpes simplex virus type 2 in vitro and in vivo. Viruses 2018, 10, 1–15. [Google Scholar] [CrossRef]
- Barnard, D.L.; Day, C.W.; Bailey, K.; Heiner, M.; Montgomery, R.; Lauridsen, L.; Chan, P.K.S.; Sidwell, R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-CoV replication in BALB/c mice. Antivir. Chem. Chemother. 2006, 17, 275–284. [Google Scholar] [CrossRef]
- Mizui, T.; Yamashina, S.; Tanida, I.; Takei, Y.; Ueno, T.; Sakamoto, N.; Ikejima, K.; Kitamura, T.; Enomoto, N.; Sakai, T.; et al. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J. Gastroenterol. 2010, 45, 195–203. [Google Scholar] [CrossRef]
- Eng, E.O.; Chew, J.S.W.; Jin, P.L.; Chua, R.C.S. In vitro inhibition of human influenza A virus replication by chloroquine. Virol. J. 2006, 3, 3–5. [Google Scholar]
- Yan, Y.; Zou, Z.; Sun, Y.; Li, X.; Xu, K.F.; Wei, Y.; Jin, N.; Jiang, C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013, 23, 300–302. [Google Scholar] [CrossRef]
- Paton, N.I.; Lee, L.; Xu, Y.; Ooi, E.E.; Cheung, Y.B.; Archuleta, S.; Wong, G.; Smith, A.W. Chloroquine for influenza prevention: A randomised, double-blind, placebo controlled trial. Lancet Infect. Dis. 2011, 11, 677–683. [Google Scholar] [CrossRef]
- De Lamballerie, X.; Boisson, V.; Reynier, J.C.; Enault, S.; Charrel, R.N.; Flahault, A.; Roques, P.; Grand, R. Le On chikungunya acute infection and chloroquine treatment. Vector Borne Zoonotic Dis. 2008, 8, 837–839. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Santhosh, S.R.; Tiwari, M.; Lakshmana Rao, P.V.; Parida, M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in Vero cells. J. Med. Virol. 2010, 82, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Farias, K.J.S.; Machado, P.R.L.; de Almeida Junior, R.F.; de Aquino, A.A.; da Fonseca, B.A.L. Chloroquine interferes with dengue-2 virus replication in U937 cells. Microbiol. Immunol. 2014, 58, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Delvecchio, R.; Higa, L.M.; Pezzuto, P.; Valadão, A.L.; Garcez, P.P.; Monteiro, F.L.; Loiola, E.C.; Dias, A.A.; Silva, F.J.M.; Aliota, M.T.; et al. Chloroquine, an endocytosis blocking agent, inhibits zika virus infection in different cell models. Viruses 2016, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Porotto, M.; Orefice, G.; Yokoyama, C.C.; Mungall, B.A.; Realubit, R.; Sganga, M.L.; Aljofan, M.; Whitt, M.; Glickman, F.; Moscona, A. Simulating Henipavirus Multicycle Replication in a Screening Assay Leads to Identification of a Promising Candidate for Therapy. J. Virol. 2009, 83, 5148–5155. [Google Scholar] [CrossRef]
- Freiberg, A.N.; Worthy, M.N.; Lee, B.; Holbrook, M.R. Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J. Gen. Virol. 2010, 91, 765–772. [Google Scholar] [CrossRef]
- Kono, M.; Tatsumi, K.; Imai, A.M.; Saito, K.; Kuriyama, T.; Shirasawa, H. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: Involvement of p38 MAPK and ERK. Antivir. Res. 2008, 77, 150–152. [Google Scholar] [CrossRef]
- Ferraris, O.; Moroso, M.; Pernet, O.; Emonet, S.; Ferrier Rembert, A.; Paranhos-Baccalà, G.; Peyrefitte, C.N. Evaluation of Crimean-Congo hemorrhagic fever virus in vitro inhibition by chloroquine and chlorpromazine, two FDA approved molecules. Antivir. Res. 2015, 118, 75–81. [Google Scholar] [CrossRef]
- Dowall, S.D.; Bosworth, A.; Watson, R.; Bewley, K.; Taylor, I.; Rayner, E.; Hunter, L.; Pearson, G.; Easterbrook, L.; Pitman, J.; et al. Chloroquine inhibited ebola virus replication in vitro but failed to protect against infection and disease in the in vivo guinea pig model. J. Gen. Virol. 2015, 96, 3484–3492. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Lecot, S.; Belouzard, S.; Dubuisson, J.; Rouillé, Y. Bovine Viral Diarrhea Virus Entry is Dependent on Clathrin-Mediated Endocytosis. J. Virol. 2005, 79, 10826–10829. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.; Belouzard, S.; Goueslain, L.; Wakita, T.; Dubuisson, J.; Wychowski, C.; Rouillé, Y. Hepatitis C Virus Entry Depends on Clathrin-Mediated Endocytosis. J. Virol. 2006, 80, 6964–6972. [Google Scholar] [CrossRef]
- Tsiang, H.; Superti, F. Ammonium chloride and chloroquine inhibit rabies virus infection in neuroblastoma cells. Arch. Virol. 1984, 81, 377–382. [Google Scholar] [CrossRef]
- Kronenberger, P.; Vrijsen, R.; Boeyé, A. Chloroquine induces empty capsid formation during poliovirus eclipse. J. Virol. 1991, 65, 7008–7011. [Google Scholar] [CrossRef]
- Romanelli, F.; Smith, K.; Hoven, A. Chloroquine and Hydroxychloroquine as Inhibitors of Human Immunodeficiency Virus (HIV-1) Activity. Curr. Pharm. Des. 2005, 10, 2643–2648. [Google Scholar] [CrossRef]
- Bishop, N.E. Examination of potential inhibitors of hepatitis A virus uncoating. Intervirology 1998, 41, 261–271. [Google Scholar] [CrossRef]
- Mehta, A.; Ouzounov, S.; Jordan, R.; Simsek, E.; Lu, X.; Moriarty, R.M.; Jacob, G.; Dwek, R.A.; Block, T.M. Imino sugars that are less toxic but more potent as antivirals, in vitro, compared with N-n-nonyl DNJ. Antivir. Chem. Chemother. 2002, 13, 299–304. [Google Scholar] [CrossRef]
- Wu, S.-F.; Lee, C.-J.; Liao, C.-L.; Dwek, R.A.; Zitzmann, N.; Lin, Y.-L. Antiviral Effects of an Iminosugar Derivative on Flavivirus Infections. J. Virol. 2002, 76, 3596–3604. [Google Scholar] [CrossRef]
- Zitzmann, N.; Mehta, A.S.; Carrouée, S.; Butters, T.D.; Platt, F.M.; McCauley, J.; Blumberg, B.S.; Dwek, R.A.; Block, T.M. Imino sugars inhibit the formation and secretion of bovine viral diarrhea virus, a pestivirus model of hepatitis C virus: Implications for the development of broad spectrum anti-hepatitis virus agents. Proc. Natl. Acad. Sci. USA 1999, 96, 11878–11882. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, E.; Whitfield, T.; Kallis, S.; Dwek, R.A.; Zitzmann, N.; Pietschmann, T.; Bartenschlager, R. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 2007, 46, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Block, T.M.; Lu, X.; Mehta, A.S.; Blumberg, B.S.; Tennant, B.; Ebling, M.; Korba, B.; Lansky, D.M.; Jacob, G.S.; Dwek, R.A. Treatment of chronic hepadnavirus infection in a woodchuck animal model with an inhibitor of protein folding and trafficking. Nat. Med. 1998, 4, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Mason, P.; Wang, L.; Norton, P.; Bourne, N.; Moriarty, R.; Mehta, A.; Despande, M.; Shah, R.; Block, T. Antiviral profiles of novel iminocyclitol compounds against bovine viral diarrhea virus, West Nile virus, dengue virus and hepatitis B virus. Antivir. Chem. Chemother. 2007, 18, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wang, L.; Ma, D.; Qu, X.; Guo, H.; Xu, X.; Mason, P.M.; Bourne, N.; Moriarty, R.; Gu, B.; et al. Novel imino sugar derivatives demonstrate potent antiviral activity against flaviviruses. Antimicrob. Agents Chemother. 2009, 53, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Pan, X.; Weidner, J.; Yu, W.; Alonzi, D.; Xu, X.; Butters, T.; Block, T.; Guo, J.T.; Chang, J. Inhibitors of endoplasmic reticulum α-glucosidases potently suppress hepatitis C virus virion assembly and release. Antimicrob. Agents Chemother. 2011, 55, 1036–1044. [Google Scholar] [CrossRef]
- Fukushi, M.; Yoshinaka, Y.; Matsuoka, Y.; Hatakeyama, S.; Ishizaka, Y.; Kirikae, T.; Sasazuki, T.; Miyoshi-Akiyama, T. Monitoring of S Protein Maturation in the Endoplasmic Reticulum by Calnexin is Important for the Infectivity of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2012, 86, 11745–11753. [Google Scholar] [CrossRef]
- Goodfellow, V.S.; Loweth, C.J.; Ravula, S.B.; Wiemann, T.; Nguyen, T.; Xu, Y.; Todd, D.E.; Sheppard, D.; Pollack, S.; Polesskaya, O.; et al. Discovery, synthesis, and characterization of an orally bioavailable, brain penetrant inhibitor of mixed lineage kinase 3. J. Med. Chem. 2013, 56, 8032–8048. [Google Scholar] [CrossRef]
- Marker, D.F.; Tremblay, M.È.; Puccini, J.M.; Barbieri, J.; Gantz Marker, M.A.; Loweth, C.J.; Chris Muly, E.; Lu, S.M.; Goodfellow, V.S.; Dewhurst, S.; et al. The new small-molecule mixed-lineage kinase 3 inhibitor URMC-099 is neuroprotective and anti-inflammatory in models of human immunodeficiency virus-associated neurocognitive disorders. J. Neurosci. 2013, 33, 9998–10010. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, D.; Dash, P.K.; Araínga, M.; Wiederin, J.L.; Haverland, N.A.; Knibbe-Hollinger, J.; Martinez-Skinner, A.; Ciborowski, P.; Goodfellow, V.S.; et al. The mixed lineage kinase-3 inhibitor URMC-099 improves therapeutic outcomes for long-acting antiretroviral therapy. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 109–122. [Google Scholar] [CrossRef]
- Xu, H.; Cheng, M.; Chi, X.; Liu, X.; Zhou, J.; Lin, T.; Yang, W. High-Throughput Screening Identifies Mixed-Lineage Kinase 3 as a Key Host Regulatory Factor in Zika Virus Infection. J. Virol. 2019, 93, 1–16. [Google Scholar] [CrossRef]
- Warren, T.K.; Jordan, R.; Lo, M.K.; Ray, A.S.; Mackman, R.L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H.C.; et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Jordan, R.; Arvey, A.; Sudhamsu, J.; Shrivastava-Ranjan, P.; Hotard, A.L.; Flint, M.; McMullan, L.K.; Siegel, D.; Clarke, M.O.; et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Leist, S.R.; Schäfer, A.; Won, J.; Brown, A.J.; Montgomery, S.A.; Hogg, A.; Babusis, D.; Clarke, M.O.; et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Graham, R.L.; Menachery, V.D.; Gralinski, L.E.; Case, J.B.; Leist, S.R.; Pyrc, K.; Feng, J.Y.; Trantcheva, I.; et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holwerda, M.; V’kovski, P.; Wider, M.; Thiel, V.; Dijkman, R. Identification of an Antiviral Compound from the Pandemic Response Box that Efficiently Inhibits SARS-CoV-2 Infection In Vitro. Microorganisms 2020, 8, 1872. https://doi.org/10.3390/microorganisms8121872
Holwerda M, V’kovski P, Wider M, Thiel V, Dijkman R. Identification of an Antiviral Compound from the Pandemic Response Box that Efficiently Inhibits SARS-CoV-2 Infection In Vitro. Microorganisms. 2020; 8(12):1872. https://doi.org/10.3390/microorganisms8121872
Chicago/Turabian StyleHolwerda, Melle, Philip V’kovski, Manon Wider, Volker Thiel, and Ronald Dijkman. 2020. "Identification of an Antiviral Compound from the Pandemic Response Box that Efficiently Inhibits SARS-CoV-2 Infection In Vitro" Microorganisms 8, no. 12: 1872. https://doi.org/10.3390/microorganisms8121872
APA StyleHolwerda, M., V’kovski, P., Wider, M., Thiel, V., & Dijkman, R. (2020). Identification of an Antiviral Compound from the Pandemic Response Box that Efficiently Inhibits SARS-CoV-2 Infection In Vitro. Microorganisms, 8(12), 1872. https://doi.org/10.3390/microorganisms8121872





