The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life
Abstract
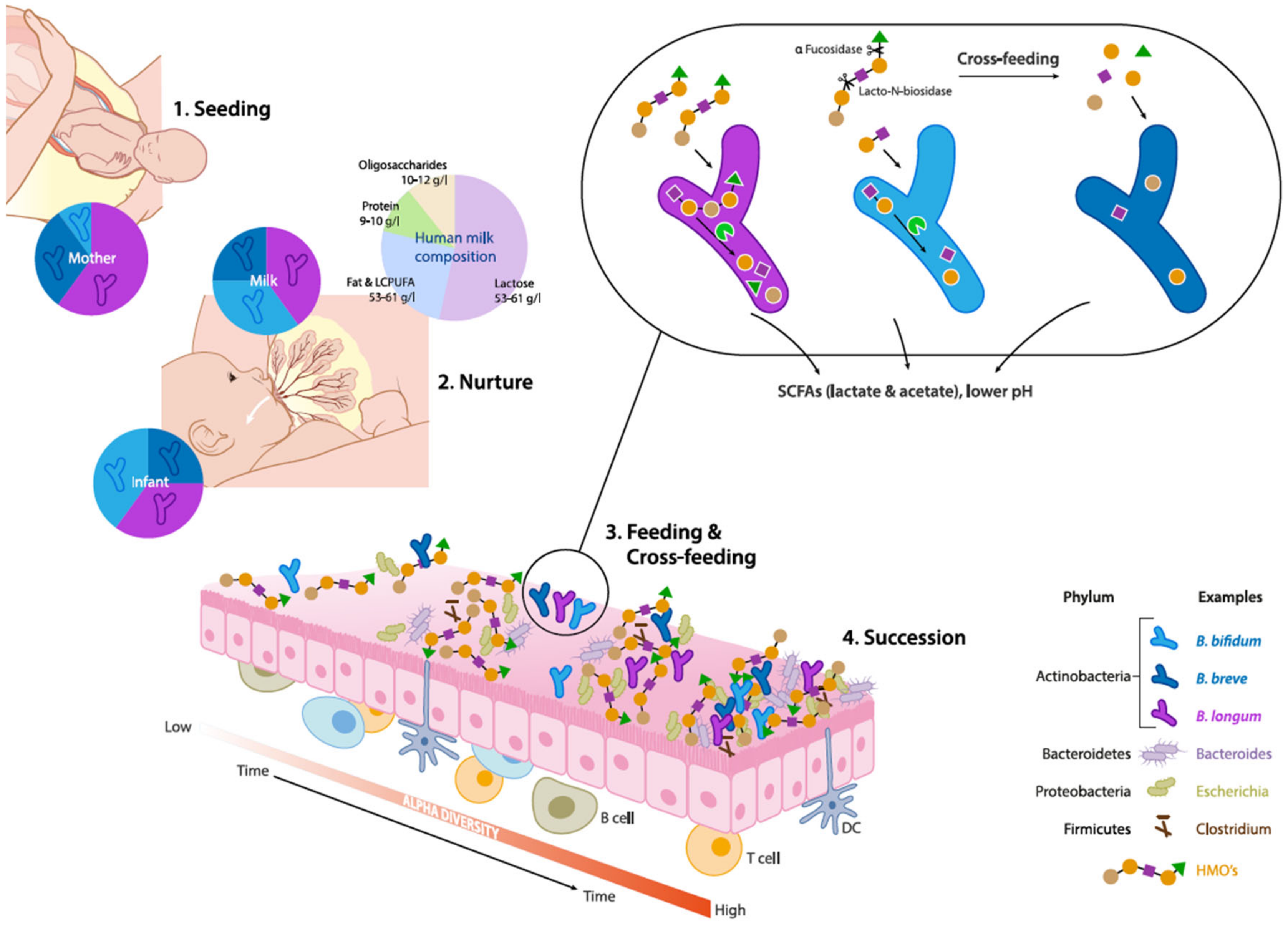
1. Introduction
2. Ecological Drivers of Acquisition and Succession of Bifidobacteria
2.1. Mode of Delivery, Antibiotics and Diet
2.2. Establishment of Bifidobacterium: A Keystone Genus
3. Evolutionary and Eco-Physiological Attributes of a Bifidogenic Milieu
3.1. Evolutionary Ecology
3.2. Genomic Features
3.3. HMO Utilization
3.4. Effect on pH and SCFA Production
4. Infant Type Bifidobacteria as Indigenous Probiotics
4.1. Comparison with Probiotic Lactobacillus Species
4.2. Beneficial Effects of Bifidobacterium Breve Strains on Infant Health
4.3. Preterm Infants
4.4. Cesarean Section
5. Allergy Development
6. Emerging Relevance of Bifidobacteria in Later Life
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wopereis:, H.; Oozeer, R.; Knipping, K.; Belzer, C.; Knol, J. The first thousand days—Intestinal microbiology of early life: Establishing a symbiosis. Pediatr. Allergy Immunol. 2014, 25, 428–438. [Google Scholar] [CrossRef]
- Houghteling, P.D.; Walker, W.A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 2015, 60, 294–307. [Google Scholar] [CrossRef]
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.e135. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.-F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef]
- Makino, H.; Martin, R.; Ishikawa, E.; Gawad, A.; Kubota, H.; Sakai, T.; Oishi, K.; Tanaka, R.; Ben-Amor, K.; Knol, J.; et al. Multilocus sequence typing of bifidobacterial strains from infant’s faeces and human milk: Are bifidobacteria being sustainably shared during breastfeeding? Benefic. Microbes 2015, 6, 563–572. [Google Scholar] [CrossRef]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Hobbs, A.J.; Mannion, C.A.; McDonald, S.W.; Brockway, M.; Tough, S.C. The impact of caesarean section on breastfeeding initiation, duration and difficulties in the first four months postpartum. BMC Pregnancy Childbirth 2016, 16, 90. [Google Scholar] [CrossRef]
- Scholtens, P.A.M.J.; Oozeer, R.; Martin, R.; Amor, K.B.; Knol, J. The early settlers: Intestinal microbiology in early life. Annu. Rev. Food Sci. Technol. 2012, 3, 425–447. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Yan, S. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019. [Google Scholar] [CrossRef]
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Lang, J.E.; Garssen, J.; Kraneveld, A.; Maitland-van der Zee, A.H. Early-life antibiotic exposure increases the risk of developing allergic symptoms later in life: A meta-analysis. Allergy 2018, 73, 971–986. [Google Scholar] [CrossRef]
- Turta, O.; Rautava, S. Antibiotics, obesity and the link to microbes—What are we doing to our children? BMC Med. 2016, 14, 57. [Google Scholar] [CrossRef]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef]
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 2019, 6. [Google Scholar] [CrossRef]
- Di Gioia, D.; Aloisio, I.; Mazzola, G.; Biavati, B. Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl. Microbiol. Biotechnol. 2014, 98, 563–577. [Google Scholar] [CrossRef]
- Martin, R.; Makino, H.; Yavuz, A.C.; Ben-Amor, K.; Roelofs, M.; Ishikawa, E.; Kubota, H.; Swinkels, S.; Sakai, T.; Oishi, K.; et al. Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Voreades, N.; Kozil, A.; Weir, T.L. Diet and the development of the human intestinal microbiome. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Paine, R.T. A note on trophic complexity and community stability. Am. Nat. 1969, 103, 91–93. [Google Scholar] [CrossRef]
- Newburg, D.S.; Morelli, L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr. Res. 2015, 77, 115–120. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Arrieta, M.-C. Patterns of early-life gut microbial colonization during human immune development: An ecological perspective. Front. Immunol. 2017, 8, 788. [Google Scholar] [CrossRef]
- Henrick, B.; Hutton, A.; Palumbo, M.; Casaburi, G.; Smilowitz, J.; Underwood, M.; Frese, S. Changes in infant fecal pH shows a population-wide, generational loss of Bifidobacterium among breastfed infants over the last century. J. Pediatric. Gastroenterol. Nutr. 2018, 66, 1073. [Google Scholar]
- Yu, Z.-T.; Chen, C.; Kling, D.E.; Liu, B.; McCoy, J.M.; Merighi, M.; Heidtman, M.; Newburg, D.S. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology 2013, 23, 169–177. [Google Scholar] [CrossRef]
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564. [Google Scholar] [CrossRef]
- Milani, C.; Turroni, F.; Duranti, S.; Lugli, G.A.; Mancabelli, L.; Ferrario, C.; van Sinderen, D.; Ventura, M. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl.Environ. Microbiol. 2015, 82, 980–991. [Google Scholar] [CrossRef]
- Ventura, M.; Turroni, F.; Lugli, G.A.; van Sinderen, D. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J. Sci. Food Agric. 2014, 94, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.B.; Sugahara, H.; Odamaki, T.; Xiao, J.Z. Different physiological properties of human-residential and non-human-residential bifidobacteria in human health. Benefic. Microbes 2017, 9, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Mahony, J.; van Sinderen, D.; Ventura, M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015, 81, 7078–7087. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Ben Amor, K.; Oozeer, R.; et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793. [Google Scholar] [CrossRef]
- Korpela, K.; Costea, P.; Coelho, L.P.; Kandels-Lewis, S.; Willemsen, G.; Boomsma, D.I.; Segata, N.; Bork, P. Selective maternal seeding and environment shape the human gut microbiome. Genome. Res. 2018, 28, 561–568. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef]
- Vaiserman, A.M.; Koliada, A.K.; Marotta, F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 2017, 35, 36–45. [Google Scholar] [CrossRef]
- Kato, K.; Odamaki, T.; Mitsuyama, E.; Sugahara, H.; Xiao, J.-z.; Osawa, R. Age-related changes in the composition of gut bifidobacterium species. Curr. Microbiol. 2017, 74, 987–995. [Google Scholar] [CrossRef]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef]
- Bottacini, F.; van Sinderen, D.; Ventura, M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem. J. 2017, 474, 4137–4152. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Bottacini, F.; Mangifesta, M.; Sanchez, B.; Viappiani, A.; Mancabelli, L.; Taminiau, B.; et al. Genomic encyclopedia of type strains of the genus bifidobacterium. Appl. Environ. Microbiol. 2014, 80, 6290–6302. [Google Scholar] [CrossRef] [PubMed]
- Meurant, G. Handbook of Milk Composition; Elsevier Science: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Zivkovic, A.M.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 4653. [Google Scholar] [CrossRef] [PubMed]
- Stahl, B.; Thurl, S.; Zeng, J.; Karas, M.; Hillenkamp, F.; Steup, M.; Sawatzki, G. Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 1994, 223, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moyano, S.; Totten, S.M.; Garrido, D.A.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl. Environ. Microbiol. 2013, 79, 6040–6049. [Google Scholar] [CrossRef] [PubMed]
- Thomson, P.; Medina, D.A.; Garrido, D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018, 75, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Hansen, M.E.; Gotoh, A.; Katoh, T.; Yoshida, K.; Odamaki, T.; Yachi, H.; Sugiyama, Y.; Kurihara, S.; Hirose, J.; et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019, 5, eaaw7696. [Google Scholar] [CrossRef]
- Garrido, D.; Barile, D.; Mills, D.A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012, 3, 415S–421S. [Google Scholar] [CrossRef]
- Zúñiga, M.; Monedero, V.; Yebra, M.J. Utilization of host-derived glycans by intestinal lactobacillus and bifidobacterium species. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Milani, C.; Lugli, G.A.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferrario, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 2016, 10, 1656. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr. Rev. 2015, 73, 426–437. [Google Scholar] [CrossRef]
- Boehm, G.; Moro, G. Structural and functional aspects of prebiotics used in infant nutrition. J. Nutr. Suppl. 2008, 138, 1818s–1828s. [Google Scholar] [CrossRef]
- Alliet, P.; Puccio, G.; Janssens, E.; Cajozzo, C.; Corsello, G.; Berger, B.; Sperisen, P.; Martin, F.-P.; Sprenger, N.; Steenhout, P. Term Infant Formula Supplemented with Human Milk Oligosaccharides (2′fucosyllactose and Lacto-Neotetraose) Shifts Stoom Microbiota and Metabolic Signatures Closer to that of Breastfed Infants. J. Pediatric Gastroenterol. Nutr. 2016, 63, S55. [Google Scholar] [CrossRef]
- Elison, E.; Vigsnaes, L.K.; Rindom Krogsgaard, L.; Rasmussen, J.; Sørensen, N.; McConnell, B.; Hennet, T.; Sommer, M.O.A.; Bytzer, P. Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br. J. Nutr. 2016, 116, 1356–1368. [Google Scholar] [CrossRef]
- Bridgman, S.L.; Azad, M.B.; Field, C.J.; Haqq, A.M.; Becker, A.B.; Mandhane, P.J.; Subbarao, P.; Turvey, S.E.; Sears, M.R.; Scott, J.A.; et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: Differences in relative versus absolute concentrations. Front. Nutr. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.T.; Lacroix, C.; Braegger, C.P.; Chassard, C. Early colonization of functional groups of microbes in the infant gut. Environ. Microbiol. 2016, 18, 2246–2258. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A.; Parrett, A.M.; Balmer, S.E.; Wharton, B.A. Faecal short chain fatty acids in breast-fed and formula-fed babies. Acta Paediatr. 1994, 83, 459–462. [Google Scholar] [CrossRef]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere 2018, 3, e00041-00018. [Google Scholar] [CrossRef]
- Orrhage, K.; Nord, C.E. Bifidobacteria and lactobacilli in human health. Drugs Exp. Clin. Res. 2000, 26, 95–111. [Google Scholar]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef]
- Wopereis, H.; Sim, K.; Shaw, A.; Warner, J.O.; Knol, J.; Kroll, J.S. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J. Allergy Clin. Immunol. 2017. [Google Scholar] [CrossRef]
- Kumari, M.; Kozyrskyj, A.L. Gut microbial metabolism defines host metabolism: an emerging perspective in obesity and allergic inflammation. Obes. Rev. 2017, 18, 18–31. [Google Scholar] [CrossRef]
- Schwab, C.; Ruscheweyh, H.-J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic Interactions of infant bifidobacteria and eubacterium hallii during L-Fucose and fucosyllactose degradation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Lee, H.; Shen, H.; Hwang, I.; Ling, H.; Yew, W.; Lee, Y.; Chang, M. Targeted Approaches for in situ gut microbiome manipulation. Genes 2018, 9, 351. [Google Scholar] [CrossRef]
- Kumar, H.; Salminen, S. Probiotics. In Encyclopedia of Food and Health; Academic Press: Oxford, UK, 2016; pp. 510–515. [Google Scholar] [CrossRef]
- Kumar, H.; Salminen, S. Probiotics: Interactions with gut microbiota and emerging trends. In Beneficial Microbes in Fermented and Functional Foods; CRC Press: Boca Raton, FL, USA, 2014; pp. 387–394. [Google Scholar] [CrossRef]
- McLaughlin, H.P.; Motherway, M.O.C.; Lakshminarayanan, B.; Stanton, C.; Paul Ross, R.; Brulc, J.; Menon, R.; O’Toole, P.W.; van Sinderen, D. Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int. J. Food Microbiol. 2015, 203, 109–121. [Google Scholar] [CrossRef]
- Nagpal, R.; Wang, S.; Ahmadi, S.; Hayes, J.; Gagliano, J.; Subashchandrabose, S.; Kitzman, D.W.; Becton, T.; Read, R.; Yadav, H. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep. 2018, 8, 12649. [Google Scholar] [CrossRef]
- Reuter, G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2001, 2, 43–53. [Google Scholar]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193. [Google Scholar] [CrossRef]
- Walter, J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 2008, 74, 4985–4996. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Thijs, C.; Vink, C.; Stelma, F.F.; Snijders, B.; Kummeling, I.; van den Brandt, P.A.; Stobberingh, E.E. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics 2006, 118, 511. [Google Scholar] [CrossRef] [PubMed]
- Nuriel-Ohayon, M.; Neuman, H.; Ziv, O.; Belogolovski, A.; Barsheshet, Y.; Bloch, N.; Uzan, A.; Lahav, R.; Peretz, A.; Frishman, S.; et al. Progesterone increases bifidobacterium relative abundance during late pregnancy. Cell Rep. 2019, 27, 730–736.e733. [Google Scholar] [CrossRef] [PubMed]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Kling Bäckhed, H.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, A.; Collado, M.C.; Garcia-Valdes, L.; Segura, M.T.; Martin-Lagos, J.A.; Anjos, T.; Marti-Romero, M.; Lopez, R.M.; Florido, J.; Campoy, C.; et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010, 104, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Mining lactobacillus and bifidobacterium for organisms with long-term gut colonization potential. Clin. Nutr. 2019. [Google Scholar] [CrossRef]
- Wong, C.B.; Odamaki, T.; Xiao, J.-z. Beneficial effects of Bifidobacterium longum subsp. longum BB536 on human health: Modulation of gut microbiome as the principal action. J. Funct. Foods 2019, 54, 506–519. [Google Scholar] [CrossRef]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229–235. [Google Scholar] [CrossRef]
- Chichlowski, M.; Shah, N.; Wampler, J.L.; Wu, S.S.; Vanderhoof, J.A. Bifidobacterium longum subspecies infantis (b. infantis) in pediatric nutrition: Current state of knowledge. Nutrients 2020, 12, 1581. [Google Scholar] [CrossRef]
- Turroni, F.; Duranti, S.; Milani, C.; Lugli, G.A.; van Sinderen, D.; Ventura, M. Bifidobacterium bifidum: A Key Member of the Early Human Gut Microbiota. Microorganisms 2019, 7. [Google Scholar] [CrossRef]
- Yaeshima, T.; Takahashi, S.; Ishibashi, N.; Shimamura, S. Identification of bifidobacteria from dairy products and evaluation of a microplate hybridization method. Int. J. Food Microbiol. 1996, 30, 303–313. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Chapter 15—Bifidobacterium breve A2—Floch, Martin, H. In The Microbiota in Gastrointestinal Pathophysiology; Ringel, Y., Walker, W.A., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 135–137. [Google Scholar] [CrossRef]
- O’Neill, I.; Schofield, Z.; Hall, L.J. Exploring the role of the microbiota member Bifidobacterium in modulating immune-linked diseases. Emerg. Top. Life Sci. 2017, 1, 333–349. [Google Scholar] [CrossRef]
- Van de Pol, M.A.; Lutter, R.; Smids, B.S.; Weersink, E.J.; van der Zee, J.S. Synbiotics reduce allergen-induced T-helper 2 response and improve peak expiratory flow in allergic asthmatics. Allergy 2011, 66, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Braga, T.D.; da Silva, G.A.; de Lira, P.I.; de Carvalho Lima, M. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: A double-blind, randomized, controlled trial. Am. J. Clin. Nutr. 2011, 93, 81–86. [Google Scholar] [CrossRef]
- Souza, D.N.; Jorge, M.T. The effect of Lactobacillus casei and Bifidobacterium breve on antibiotic-associated diarrhea treatment: randomized double-blind clinical trial. Rev. Soc. Bras. Med. Trop. 2012, 45, 112–116. [Google Scholar] [CrossRef]
- Wada, M.; Nagata, S.; Saito, M.; Shimizu, T.; Yamashiro, Y.; Matsuki, T.; Asahara, T.; Nomoto, K. Effects of the enteral administration of Bifidobacterium breve on patients undergoing chemotherapy for pediatric malignancies. Supportive Care Cancer 2010, 18, 751–759. [Google Scholar] [CrossRef]
- Klemenak, M.; Dolinsek, J.; Langerholc, T.; Di Gioia, D.; Micetic-Turk, D. Administration of Bifidobacterium breve decreases the production of TNF-alpha in children with celiac disease. Dig. Dis. Sci. 2015, 60, 3386–3392. [Google Scholar] [CrossRef]
- Abrahamse-Berkeveld, M.; Alles, M.; Franke-Beckmann, E.; Helm, K.; Knecht, R.; Köllges, R.; Sandner, B.; Knol, J.; Ben Amor, K.; Bufe, A. Infant formula containing galacto-And fructo-oligosaccharides and Bifidobacterium breve M-16V supports adequate growth and tolerance in healthy infants in a randomised, controlled, double-blind, prospective, multicentre study. J. Nutr. Sci. 2016, 5. [Google Scholar] [CrossRef]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A synbiotic-containing amino-acid-based formula improves gut microbiota in non-IgE-mediated allergic infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef]
- Fanning, S.; Hall, L.J.; Cronin, M.; Zomer, A.; MacSharry, J.; Goulding, D.; Connell Motherway, M.; Shanahan, F.; Nally, K.; Dougan, G.; et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. USA 2012, 109, 2108. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Tsilingiri, K.; Rescigno, M. Postbiotics: what else? Benef. Microbes. 2013, 4, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Mullie, C.; Yazourh, A.; Thibault, H.; Odou, M.F.; Singer, E.; Kalach, N.; Kremp, O.; Romond, M.B. Increased poliovirus-specific intestinal antibody response coincides with promotion of Bifidobacterium longum-infantis and Bifidobacterium breve in infants: A randomized, double-blind, placebo-controlled trial. Pediatr. Res. 2004, 56, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Thibault, H.; Aubert-Jacquin, C.; Goulet, O. Effects of long-term consumption of a fermented infant formula (with Bifidobacterium breve c50 and Streptococcus thermophilus 065) on acute diarrhea in healthy infants. J. Pediatr. Gastroenterol. Nutr. 2004, 39, 147–152. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Ludwig, T.; Bouritius, H.; Alliet, P.; Forde, D.; Peeters, S.; Huet, F.; Hourihane, J. Randomised controlled trial demonstrates that fermented infant formula with short-chain galacto-oligosaccharides and long-chain fructo-oligosaccharides reduces the incidence of infantile colic. Acta Paediatr. 2017, 106, 1150–1158. [Google Scholar] [CrossRef]
- Huet, F.; Abrahamse-Berkeveld, M.; Tims, S.; Simeoni, U.; Beley, G.; Savagner, C.; Vandenplas, Y.; Hourihane, J.O.B. Partly fermented infant formulae with specific oligosaccharides support adequate infant growth and are well-tolerated. J. Pediatric Gastroenterol. Nutr. 2016, 63, e43–e53. [Google Scholar] [CrossRef]
- Chawanpaiboon, S.; Vogel, J.P.; Moller, A.-B.; Lumbiganon, P.; Petzold, M.; Hogan, D.; Landoulsi, S.; Jampathong, N.; Kongwattanakul, K.; Laopaiboon, M.; et al. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob. Health 2019, 7, e37–e46. [Google Scholar] [CrossRef]
- Arboleya, S.; Ang, L.; Margolles, A.; Yiyuan, L.; Dongya, Z.; Liang, X.; Solís, G.; Fernández, N.; Clara, G.; Gueimonde, M.J.A. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe 2012, 18, 378–380. [Google Scholar] [CrossRef]
- Barrett, E.; Kerr, C.; Murphy, K.; O’Sullivan, O.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.P.; O’Toole, P.W.; Cotter, P.D.; Fitzgerald, G.F.; et al. The individual-specific and diverse nature of the preterm infant microbiota. Arch. Dis. Child. Fetal Neonatal Ed. 2013, 98, F334–F340. [Google Scholar] [CrossRef]
- Gibson, M.K.; Wang, B.; Ahmadi, S.; Burnham, C.-A.D.; Tarr, P.I.; Warner, B.B.; Dantas, G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 2016, 1, 16024. [Google Scholar] [CrossRef]
- Schwiertz, A.; Gruhl, B.; Lobnitz, M.; Michel, P.; Radke, M.; Blaut, M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr. Res. 2003, 54, 393–399. [Google Scholar] [CrossRef]
- Patel, A.L.; Mutlu, E.A.; Sun, Y.; Koenig, L.; Green, S.; Jakubowicz, A.; Mryan, J.; Engen, P.; Fogg, L.; Chen, A.L.; et al. Longitudinal survey of microbiota in hospitalized preterm very-low-birth-weight infants. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 292–303. [Google Scholar] [CrossRef]
- Kurath-Koller, S.; Neumann, C.; Moissl-Eichinger, C.; Kraschl, R.; Kanduth, C.; Hopfer, B.; Pausan, M.R.; Urlesberger, B.; Resch, B. Hospital regimens including probiotics guide the individual development of the gut microbiome of very low birth weight infants in the first two weeks of life. Nutrients 2020, 12, 1256. [Google Scholar] [CrossRef]
- Zwittink, R.D.; Renes, I.B.; van Lingen, R.A.; van Zoeren-Grobben, D.; Konstanti, P.; Norbruis, O.F.; Martin, R.; Groot Jebbink, L.J.M.; Knol, J.; Belzer, C. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 475–483. [Google Scholar] [CrossRef]
- Patole, S.K.; Rao, S.C.; Keil, A.D.; Nathan, E.A.; Doherty, D.A.; Simmer, K.N. Benefits of Bifidobacterium breve M-16V Supplementation in Preterm Neonates - A Retrospective Cohort Study. PLoS ONE 2016, 11, e0150775. [Google Scholar] [CrossRef]
- Costeloe, K.; Hardy, P.; Juszczak, E.; Wilks, M.; Millar, M.R.; Probiotics in Preterm Infants Study Collaborative, G. Bifidobacterium breve BBG-001 in very preterm infants: A randomised controlled phase 3 trial. Lancet 2016, 387, 649–660. [Google Scholar] [CrossRef]
- Athalye-Jape, G.; Rao, S.; Simmer, K.; Patole, S. Bifidobacterium breve M-16V as a probiotic for preterm infants: A strain-specific systematic review. JPEN J. Parenter. Enteral. Nutr. 2017. [Google Scholar] [CrossRef]
- Boerma, T.; Ronsmans, C.; Melesse, D.Y.; Barros, A.J.D.; Barros, F.C.; Juan, L.; Moller, A.-B.; Say, L.; Hosseinpoor, A.R.; Yi, M.; et al. Global epidemiology of use of and disparities in caesarean sections. Lancet 2018, 392, 1341–1348. [Google Scholar] [CrossRef]
- Jessica, P. Caesarean section and children’s health: A quasi-experimental design. Popul. Stud. 2019. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Wampach, L.; Heintz-Buschart, A.; Fritz, J.V.; Ramiro-Garcia, J.; Habier, J.; Herold, M.; Narayanasamy, S.; Kaysen, A.; Hogan, A.H.; Bindl, L.; et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 2018, 9, 5091. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef]
- Dierikx, T.H.; Berkhout, D.J.C.; Visser, L.; Benninga, M.A.; Roeselers, G.; de Boer, N.K.H.; de Vries, J.I.P.; de Meij, T.G.J. The influence of timing of Maternal administration of Antibiotics during cesarean section on the intestinal Microbial colonization in Infants (MAMI-trial): Study protocol for a randomised controlled trial. Trials 2019, 20, 479. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vina, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250–253. [Google Scholar] [CrossRef]
- Stinson, L.F.; Payne, M.S.; Keelan, J.A. A Critical review of the bacterial baptism hypothesis and the impact of cesarean delivery on the infant microbiome. Front. Med. 2018, 5. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sakata, S.; Kalliomaki, M.; Isolauri, E.; Benno, Y.; Salminen, S. Effect of maternal consumption of lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 166–170. [Google Scholar] [CrossRef]
- Barrett, E.; Deshpandey, A.K.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; O’Sullivan, L.; Watkins, C.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; et al. The neonatal gut harbours distinct bifidobacterial strains. Arch. Dis. Child. Fetal. Neonatal. Ed. 2015, 100, F405–F410. [Google Scholar] [CrossRef]
- Chua, M.C.; Ben-Amor, K.; Lay, C.; Neo, A.G.E.; Chiang, W.C.; Rao, R.; Chew, C.; Chaithongwongwatthana, S.; Khemapech, N.; Knol, J.; et al. Effect of synbiotic on the gut microbiota of cesarean delivered infants: A randomized, double-blind, multicenter study. J. Pediatric Gastroenterol. Nutr. 2017, 65, 102–106. [Google Scholar] [CrossRef]
- Akdis, C.A.; Agache, I. Global Atlas of Allergy; European Academy of Allergy and Clinical Immunology: Zurich, Switzerland, 2014. [Google Scholar]
- Kalliomaki, M.; Kirjavainen, P.; Eerola, E.; Kero, P.; Salminen, S.; Isolauri, E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 2001, 107, 129–134. [Google Scholar] [CrossRef]
- Akay, H.K.; Bahar Tokman, H.; Hatipoglu, N.; Hatipoglu, H.; Siraneci, R.; Demirci, M.; Borsa, B.A.; Yuksel, P.; Karakullukcu, A.; Kangaba, A.A.; et al. The relationship between bifidobacteria and allergic asthma and/or allergic dermatitis: A prospective study of 0–3 years-old children in Turkey. Anaerobe 2014, 28, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Melli, L.C.; do Carmo-Rodrigues, M.S.; Araujo-Filho, H.B.; Sole, D.; de Morais, M.B. Intestinal microbiota and allergic diseases: A systematic review. Allergol. Immunopathol. 2016, 44, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Boyle, R.J.; Licciardi, P.V.; Oppedisano, F.; Lahtinen, S.; Robins‐Browne, R.M.; Tang, M.L. Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr. Allergy Immunol. 2016, 27, 838–846. [Google Scholar] [CrossRef]
- Van der Aa, L.; Van Aalderen, W.; Heymans, H.; Henk Sillevis Smitt, J.; Nauta, A.; Knippels, L.; Ben Amor, K.; Sprikkelman, A. Synbiotics prevent asthma-like symptoms in infants with atopic dermatitis. Allergy 2011, 66, 170–177. [Google Scholar] [CrossRef]
- Van der Aa, L.B.; Heymans, H.S.; van Aalderen, W.M.; Sillevis Smitt, J.H.; Knol, J.; Ben Amor, K.; Goossens, D.A.; Sprikkelman, A.B. Effect of a new synbiotic mixture on atopic dermatitis in infants: A randomized-controlled trial. Clin. Exp. Allergy 2010, 40, 795–804. [Google Scholar] [CrossRef]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Margolles, A.; Sánchez, B. Proteinaceous Molecules Mediating Bifidobacterium-Host Interactions. Front. Microbiol. 2016, 7, 1193. [Google Scholar] [CrossRef]
- Meng, D.; Zhu, W.; Ganguli, K.; Shi, H.N.; Walker, W.A. Anti-inflammatory effects of Bifidobacterium longum subsp infantis secretions on fetal human enterocytes are mediated by TLR-4 receptors. Am. J. Physiol. Gastrointest Liver Physiol. 2016, 311, G744–G753. [Google Scholar] [CrossRef]
- Castro-Bravo, N.; Wells, J.M.; Margolles, A.; Ruas-Madiedo, P. Interactions of Surface exopolysaccharides from bifidobacterium and lactobacillus within the intestinal environment. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Kado, Y.; Takada, T.; Matsumoto, K.; Tanaka, R.J.A.E.M. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 167–173. [Google Scholar] [CrossRef]
- Chaplin, A.; Brzhozovskii, A.; Parfenova, T.; Kafarskaia, L.; Volodin, N.; Shkoporov, A.; Ilina, E.; Efimov, B.A. Species diversity of bifidobacteria in the intestinal microbiota studied using MALDI-TOF mass-spectrometry. Ann. Russ. Acad. Med Sci. 2015, 70, 435–440. [Google Scholar] [CrossRef]
- Duranti, S.; Milani, C.; Lugli, G.A.; Mancabelli, L.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Sánchez, B.; Margolles, A.; et al. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 2016, 6, 23971. [Google Scholar] [CrossRef]
- Duranti, S.; Turroni, F.; Lugli, G.A.; Milani, C.; Viappiani, A.; Mangifesta, M.; Gioiosa, L.; Palanza, P.; van Sinderen, D.; Ventura, M.J.A.E.M. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl. Environ. Microbiol. 2014, 80, 6080–6090. [Google Scholar] [CrossRef]
- Gueimonde, M.; Ouwehand, A.; Pitkälä, K.; Strandberg, T.; Finne-Soveri, H.; Salminen, S. Fecal bifidobacterium levels in elderly nursing home patients. Biosci. Microflora 2010, 29, 111–113. [Google Scholar] [CrossRef]
- Mitsuoka, T. Intestinal flora and aging. Nutr. Rev. Wash. 1992, 50, 438–446. [Google Scholar] [CrossRef]
- Hopkins, M.; Macfarlane, G.T. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 2002, 51, 448–454. [Google Scholar] [CrossRef]
- Salazar, N.; López, P.; Valdés, L.; Margolles, A.; Suárez, A.; Patterson, A.M.; Cuervo, A.; Reyes-Gavilán, C.G.d.l.; Ruas-Madiedo, P.; Gonzalez, S.; et al. Microbial targets for the development of functional foods accordingly with nutritional and immune parameters altered in the elderly. J. Am. Coll. Nutr. 2013, 32, 399–406. [Google Scholar] [CrossRef]
- Hao, Y.; Huang, D.; Guo, H.; Xiao, M.; An, H.; Zhao, L.; Zuo, F.; Zhang, B.; Hu, S.; Song, S.; et al. Complete Genome Sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy chinese centenarian. J. Bacteriol. 2011, 193, 787–788. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, L.; Ren, F.; Sun, E.; Zhang, M.; Guo, H. Complete genome sequence of Bifidobacterium adolesentis BBMN23, a probiotic strain from healthy centenarian. J. Biotechnol. 2015, 198, 44–45. [Google Scholar] [CrossRef]
- Duggal, N.A. Reversing the immune ageing clock: Lifestyle modifications and pharmacological interventions. Biogerontology 2018, 19, 481–496. [Google Scholar] [CrossRef]
- Maneerat, S.; Lehtinen, M.J.; Childs, C.E.; Forssten, S.D.; Alhoniemi, E.; Tiphaine, M.; Yaqoob, P.; Ouwehand, A.C.; Rastall, R.A. Consumption of Bifidobacterium lactis Bi-07 by healthy elderly adults enhances phagocytic activity of monocytes and granulocytes. J. Nutr. Sci. 2013, 2. [Google Scholar] [CrossRef]
- Pitkala, K.H.; Strandberg, T.E.; Finne Soveri, U.H.; Ouwehand, A.C.; Poussa, T.; Salminen, S. Fermented cereal with specific bifidobacteria normalizes bowel movements in elderly nursing home residents. A randomized, controlled trial. J. Nutr. Health Aging 2007, 11, 305–311. [Google Scholar]
- Epstein, R.H. Get Me Out: A History of Childbirth from the Garden of Eden to the Sperm Bank; WW Norton & Company: New York, NY, USA, 2011. [Google Scholar]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, H.; Collado, M.C.; Wopereis, H.; Salminen, S.; Knol, J.; Roeselers, G. The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life. Microorganisms 2020, 8, 1855. https://doi.org/10.3390/microorganisms8121855
Kumar H, Collado MC, Wopereis H, Salminen S, Knol J, Roeselers G. The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life. Microorganisms. 2020; 8(12):1855. https://doi.org/10.3390/microorganisms8121855
Chicago/Turabian StyleKumar, Himanshu, Maria Carmen Collado, Harm Wopereis, Seppo Salminen, Jan Knol, and Guus Roeselers. 2020. "The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life" Microorganisms 8, no. 12: 1855. https://doi.org/10.3390/microorganisms8121855
APA StyleKumar, H., Collado, M. C., Wopereis, H., Salminen, S., Knol, J., & Roeselers, G. (2020). The Bifidogenic Effect Revisited—Ecology and Health Perspectives of Bifidobacterial Colonization in Early Life. Microorganisms, 8(12), 1855. https://doi.org/10.3390/microorganisms8121855




