Microbial Shifts Following Five Years of Cover Cropping and Tillage Practices in Fertile Agroecosystems
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site Description and Management Practices
2.2. Soil and Biomass Sampling and Procedures
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. Soil and Cover Crop Biomass Properties
3.2. Overall Characterization of the Soil Microbiome
3.3. OTUs Responses to Cover Crop Rotations and Tillage Treatments
3.3.1. Bacteria
3.3.2. Fungi and Archaea
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130164. [Google Scholar] [CrossRef] [PubMed]
- Illinois Environmental Protection Agency, Illinois Department of Agriculture, University of Illinois Extension. Illinois Nutrient Loss Reduction Strategy (INLRS). 2015. Available online: https://www2.illinois.gov/epa/Documents/iepa/water-quality/watershed-management/nlrs/nlrs-final-revised-083115.pdf (accessed on 1 November 2020).
- Pennino, M.J.; Compton, J.E.; Leibowitz, S.G. Trends in drinking water nitrate violations across the United States. Environ. Sci. Technol. 2017, 51, 13450–13460. [Google Scholar] [CrossRef] [PubMed]
- Villamil, M.B.; Bollero, G.A.; Darmody, R.G.; Simmons, F.W.; Bullock, D.G. No-till corn/soybean systems including winter cover crops. Soil Sci. Soc. Am. J. 2006, 70, 1936–1944. [Google Scholar] [CrossRef]
- Villamil, M.B.; Miguez, F.E.; Bollero, G.A. Multivariate analysis and visualization of soil quality data for no-till systems. J. Environ. Qual. 2008, 37, 2063–2069. [Google Scholar] [CrossRef]
- Daryanto, S.; Fu, B.J.; Wang, L.X.; Jacinthe, P.A.; Zhao, W.W. Quantitative synthesis on the ecosystem services of cover crops. Earth Sci. Rev. 2018, 185, 357–373. [Google Scholar] [CrossRef]
- Quemada, M.; Baranski, M.; Nobel-de Lange, M.N.J.; Vallejo, A.; Cooper, J.M. Meta-analysis of strategies to control nitrate leaching in irrigated agricultural systems and their effects on crop yield. Agric. Ecosyst. Environ. 2013, 174, 1–10. [Google Scholar] [CrossRef]
- Acuña, J.C.M.; Villamil, M.B. Short-term effects of cover crops and compaction on soil properties and soybean production in Illinois. Agron. J. 2014, 106, 860–870. [Google Scholar] [CrossRef]
- Behnke, G.D.; Villamil, M.B. Cover crop rotations affect greenhouse gas emissions and crop production in Illinois, USA. Field Crops. Res. 2019, 241, 107580. [Google Scholar] [CrossRef]
- Kaspar, T.C.; Jaynes, D.B.; Parkin, T.B.; Moorman, T.B.; Singer, J.W. Effectiveness of oat and rye cover crops in reducing nitrate losses in drainage water. Agric. Water Manag. 2012, 110, 25–33. [Google Scholar] [CrossRef]
- Tonitto, C.; David, M.B.; Drinkwater, L.E. Replacing bare fallows with cover crops in fertilizer-intensive cropping systems: A meta-analysis of crop yield and n dynamics. Agric. Ecosyst. Environ. 2006, 112, 58–72. [Google Scholar] [CrossRef]
- Behnke, G.D.; Kim, N.; Villamil, M.B. Agronomic assessment of cover cropping and tillage practices across environments. Agron. J. 2020, 1–16. [Google Scholar] [CrossRef]
- Dozier, I.A.; Behnke, G.D.; Davis, A.S.; Nafziger, E.D.; Villamil, M.B. Tillage and cover cropping effects on soil properties and crop production in illinois. Agron. J. 2017, 109, 1261–1270. [Google Scholar] [CrossRef]
- Pittelkow, C.M.; Linquist, B.A.; Lundy, M.E.; Liang, X.; Van Groenigen, K.J.; Lee, J.; Van Gestel, N.; Six, J.; Venterea, R.T.; Van Kessel, C. When does no-till yield more? A global meta-analysis. Field Crops Res. 2015, 183, 156–168. [Google Scholar] [CrossRef]
- Kibet, L.C.; Blanco-Canqui, H.; Jasa, P. Long-term tillage impacts on soil organic matter components and related properties on a typic Argiudoll. Soil Tillage Res. 2016, 155, 78–84. [Google Scholar] [CrossRef]
- Sainju, U.M.; Singh, B.P.; Whitehead, W.F. Long-term effects of tillage, cover crops, and nitrogen fertilization on organic carbon and nitrogen concentrations in sandy loam soils in Georgia, USA. Soil Tillage Res. 2002, 63, 167–179. [Google Scholar] [CrossRef]
- Singh, G.; Williard, K.W.J.; Schoonover, J.E. Cover crops and tillage influence on nitrogen dynamics in plant-soil-water pools. Soil Sci. Soc. Am. J. 2018, 82, 1572–1582. [Google Scholar] [CrossRef]
- Frasier, I.; Noellemeyer, E.; Figuerola, E.; Erijman, L.; Permingeat, H.; Quiroga, A. High quality residues from cover crops favor changes in microbial community and enhance C and N sequestration. Glob. Ecol. Conserv. 2016, 6, 242–256. [Google Scholar] [CrossRef]
- Hallama, M.; Pekrun, C.; Lambers, H.; Kandeler, E. Hidden miners—The roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant Soil 2019, 434, 7–45. [Google Scholar] [CrossRef]
- Hirsch, P.R.; Mauchline, T.H. Chapter Two—The Importance of the Microbial N Cycle in Soil for Crop Plant Nutrition; Sariaslani, S., Gadd, G.M., Eds.; Adv. Appl. Microbiol. 93; Academic Press: Cambridge, MA, USA, 2015; pp. 45–71. [Google Scholar]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef]
- Kim, N.; Zabaloy, M.C.; Guan, K.Y.; Villamil, M.B. Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol. Biochem. 2020, 142, 107701. [Google Scholar] [CrossRef]
- Zuber, S.M.; Villamil, M.B. Meta-analysis approach to assess effect of tillage on microbial biomass and enzyme activities. Soil Biol. Biochem. 2016, 97, 176–187. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, P.; Riggins, C.W.; Zabaloy, M.C.; Rodríguez-Zas, S.; Villamil, M.B. Long-term n fertilization decreased diversity and altered the composition of soil bacterial and archaeal communities. Agronomy 2019, 9, 574. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue—Bacterial families and genera: A metagenomic approach. Ecol. Indic. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Srour, A.; Ammar, H.A.; Subedi, A.; Pimentel, M.; Cook, R.L.; Bond, J.; Fakhoury, A.M. Microbial communities associated with long-term tillage and fertility treatments in a corn-soybean cropping system. Front. Microbiol. 2020, 11, 1363. [Google Scholar] [CrossRef]
- Calle, M.L. Statistical analysis of metagenomics data. Genom. Inform. 2019, 17, e6. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhou, G.; Rees, R.M.; Cao, W. Green manuring inhibits nitrification in a typical paddy soil by changing the contributions of ammonia-oxidizing archaea and bacteria. Appl. Soil Ecol. 2020, 156, 103698. [Google Scholar] [CrossRef]
- Alahmad, A.; Decocq, G.; Spicher, F.; Kheirbeik, L.; Kobaissi, A.; Tetu, T.; Dubois, F.; Duclercq, J. Cover crops in arable lands increase functional complementarity and redundancy of bacterial communities. J. Appl. Ecol. 2019, 56, 651–664. [Google Scholar] [CrossRef]
- Romdhane, S.; Spor, A.; Busset, H.; Falchetto, L.; Martin, J.; Bizouard, F.; Bru, D.; Breuil, M.-C.; Philippot, L.; Cordeau, S. Cover crop management practices rather than composition of cover crop mixtures affect bacterial communities in no-till agroecosystems. Front. Microbiol. 2019, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, Z.; Gong, Q.; Zhai, B.; Li, Z. Responses of fungal–bacterial community and network to organic inputs vary among different spatial habitats in soil. Soil Biol. Biochem. 2018, 125, 54–63. [Google Scholar] [CrossRef]
- Pascault, N.; Ranjard, L.; Kaisermann, A.; Bachar, D.; Christen, R.; Terrat, S.; Mathieu, O.; Lévêque, J.; Mougel, C.; Henault, C.; et al. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 2013, 16, 810–822. [Google Scholar] [CrossRef]
- Sharma-Poudyal, D.; Schlatter, D.; Yin, C.; Hulbert, S.; Paulitz, T. Long-term no-till: A major driver of fungal communities in dryland wheat cropping systems. PLoS ONE 2017, 12, e0184611. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhao, Z.; Gong, Q.; Zhai, B.; Li, Z. Effects of cover crop in an apple orchard on microbial community composition, networks, and potential genes involved with degradation of crop residues in soil. Biol. Fertil. Soils 2018, 54, 743–759. [Google Scholar] [CrossRef]
- Official Soil Series Descriptions. NRCS USDA. 2019. Available online: https://www.nrcs.usda.gov/wps/portal/nrcs/detail/soils/home/?cid=nrcs142p2_053587 (accessed on 12 June 2020).
- Villamil, M.B.; Nafziger, E.D. Agronomic and Environmental Assessment of Cover Crops Project Data. Mendeley Data. 2019. Available online: https://doi.org/10.17632/7rn62y2cps.1 (accessed on 1 November 2020).
- Brown, J.R. Recommended Chemical Soil Test Procedures for the North Central Region (No. 1001); University of Missouri: Columbia, MO, USA, 1998. [Google Scholar]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative pcr assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Colman, D.R.; Thomas, R.; Maas, K.R.; Takacs-Vesbach, C.D. Detection and analysis of elusive members of a novel and diverse archaeal community within a thermal spring streamer consortium. Extremophiles 2015, 19, 307–313. [Google Scholar] [CrossRef]
- Crawford, J.W.; Deacon, L.; Grinev, D.; Harris, J.A.; Ritz, K.; Singh, B.K.; Young, I. Microbial diversity affects self-organization of the soil microbe system with consequences for function. J. R. Soc. Interface 2012, 9, 1302–1310. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.; Dillon, M.; Bokulich, N.; Abnet, C.; Al-Ghalith, G.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16s rrna Gene Analysis with qiime2; Beiko, R., Hsiao, W.J.P., Eds.; Microbiome analysis. 1849; Humana Press: New York, NY, USA, 2018; pp. 113–129. [Google Scholar]
- Li, X.; Nair, A.; Wang, S.; Wang, L. Quality Control of rna-seq Experiments; Picardi, E., Ed.; RNA Bioinformatics; Humana Press: New York, NY, USA, 2015; pp. 137–146. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from Illumina amplicon data. Nat. Method 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. Mafft-dash: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of mafft for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 1 December 2019).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- SAS Institute Inc. Jmp 14 Predictive and Specialized Modeling; SAS Institute: Cary, NC, USA, 2018; 464p. [Google Scholar]
- Tracy, B.; Coopersmith, C.; Silverman, M.; Fassler, M.; McCarty, E.; Gelbard, R. 1753: Bootstrap forest model predicts neurostimulant therapy after severe traumatic brain injury. Crit. Care Med. 2020, 48, 851. [Google Scholar] [CrossRef]
- Vlasova-St Louis, I.; Chang, C.C.; Shahid, S.; French, M.A.; Bohjanen, P.R. Transcriptomic predictors of paradoxical cryptococcosis-associated immune reconstitution inflammatory syndrome. Open Forum Infect. Dis. 2018, 5, ofy157. [Google Scholar] [CrossRef]
- Tabachnick, B.G.; Fidell, L.S.; Ullman, J.B. Using Multivariate Statistics; Pearson: Boston, MA, USA, 2007. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; Wiley StatsRef: Statistics Reference Online. 2017, pp. 1–15. Available online: https://doi.org/10.1002/9781118445112.stat07841 (accessed on 12 June 2020).
- Storey, J.D. The positive false discovery rate: A bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Necpalova, M.; Anex, R.P.; Kravchenko, A.N.; Abendroth, L.J.; Del Grosso, S.J.; Dick, W.A.; Helmers, M.J.; Herzmann, D.; Lauer, J.G.; Nafziger, E.D.; et al. What does it take to detect a change in soil carbon stock? A regional comparison of minimum detectable difference and experiment duration in the north central united states. J. Soil Water Conserv. 2014, 69, 517–531. [Google Scholar] [CrossRef]
- Basche, A.D.; Miguez, F.E.; Kaspar, T.C.; Castellano, M.J. Do cover crops increase or decrease nitrous oxide emissions? A meta-analysis. J. Soil Water Conserv. 2014, 69, 471–482. [Google Scholar] [CrossRef]
- Yan, L.; Xu, X.; Xia, J. Different impacts of external ammonium and nitrate addition on plant growth in terrestrial ecosystems: A meta-analysis. Sci. Total Environ. 2019, 686, 1010–1018. [Google Scholar] [CrossRef]
- Chen, H.; Dai, Z.; Veach, A.M.; Zheng, J.; Xu, J.; Schadt, C.W. Global meta-analyses show that conservation tillage practices promote soil fungal and bacterial biomass. Agric. Ecosyst. Environ. 2020, 293, 106841. [Google Scholar] [CrossRef]
- Qi, G.; Chen, S.; Ke, L.; Ma, G.; Zhao, X. Cover crops restore declining soil properties and suppress bacterial wilt by regulating rhizosphere bacterial communities and improving soil nutrient contents. Microbiol. Res. 2020, 238, 126505. [Google Scholar] [CrossRef]
- Jian, J.; Lester, B.J.; Du, X.; Reiter, M.S.; Stewart, R.D. A calculator to quantify cover crop effects on soil health and productivity. Soil Tillage Res. 2020, 199, 104575. [Google Scholar] [CrossRef]
- Tian, Y.; Han, C.; Hu, J.; Zhao, J.; Zhang, C.; Guo, X.; Wang, X.; Xiang, W. Cellulomonas rhizosphaerae sp. Nov., a novel actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Singleton, D.R.; Furlong, M.A.; Peacock, A.D.; White, D.C.; Coleman, D.C.; Whitman, W.B. Solirubrobacter pauli gen. Nov., sp. Nov., a mesophilic bacterium within the rubrobacteridae related to common soil clones. Int. J. Syst. Evol. Microbiol. 2003, 53, 485–490. [Google Scholar] [CrossRef]
- Yuan, N.; Zeng, Y.; Feng, H.; Yu, Z.; Huang, Y. Altererythrobacter xixiisoli sp. Nov., isolated from wetland soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 3655–3659. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Birtles, R.J.; Mallet, M.-N.; Raoult, D. Massilia timonae; gen. Nov., sp. Nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 1998, 36, 2847. [Google Scholar] [CrossRef]
- Kämpfer, P. Order i. Sphingobacteriales ord. Nov. Bergey’s Manual Trust; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; 330p. [Google Scholar]
- Yabuuchi, E.; Kaneko, T.; Yano, I.; Moss, C.W.; Miyoshi, N. Sphingobacterium gen. Nov., sphingobacterium spiritivorum comb. Nov., sphingobacterium multivorum comb. Nov., sphingobacterium mizutae sp. Nov., and flavobacterium indologenes sp. Nov.: Glucose-nonfermenting gram-negative rods in cdc groups iik-2 and iib. Int. J. Syst. Evol. Microbiol. 1983, 33, 580–598. [Google Scholar] [CrossRef]
- Cavaletti, L.; Monciardini, P.; Bamonte, R.; Schumann, P.; Rohde, M.; Sosio, M.; Donadio, S. New lineage of filamentous, spore-forming, gram-positive bacteria from soil. Appl. Environ. Microbiol. 2006, 72, 4360. [Google Scholar] [CrossRef]
- Yabe, S.; Sakai, Y.; Abe, K.; Yokota, A. Diversity of Ktedonobacteria with actinomycetes-like morphology in terrestrial environments. Microbes Environ. 2017, 32, 61–70. [Google Scholar] [CrossRef]
- Drost, S.M.; Rutgers, M.; Wouterse, M.; de Boer, W.; Bodelier, P.L.E. Decomposition of mixtures of cover crop residues increases microbial functional diversity. Geoderma 2020, 361, 11. [Google Scholar] [CrossRef]
- Balota, E.L.; Calegari, A.; Nakatani, A.S.; Coyne, M.S. Benefits of winter cover crops and no-tillage for microbial parameters in a brazilian Oxisol: A long-term study. Agric. Ecosyst. Environ. 2014, 197, 31–40. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.-J.; Lee, K.; Song, J.; Cho, J.-C. Puniceicoccus vermicola gen. Nov., sp. Nov., a novel marine bacterium, and description of puniceicoccaceae fam. Nov., puniceicoccales ord. Nov., opitutaceae fam. Nov., opitutales ord. Nov. and opitutae classis nov. In the phylum ‘Verrucomicrobia’. Int. J. Syst. Evol. Microbiol. 2007, 57, 532–537. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, S.J.; Awata, T.; Nierychlo, M.; Albertsen, M.; Kindaichi, T.; Nielsen, P.H. Characterization of the in situ ecophysiology of novel phylotypes in nutrient removal activated sludge treatment plants. PLoS ONE 2015, 10, e0136424. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, Y.; Wang, Y.; Chin, F.Y.L.; Zhang, T. Cellular adhesiveness and cellulolytic capacity in anaerolineae revealed by omics-based genome interpretation. Biotechnol. Biofuels 2016, 9, 111. [Google Scholar] [CrossRef] [PubMed]
- Fawaz, M. Revealing the Ecological Role of Gemmatimonadetes through Cultivation and Molecular Analysis of Agricultural Soils; University of Tennessee: Knoxville, TN, USA, 2013. [Google Scholar]
- Fernandez, A.L.; Sheaffer, C.C.; Wyse, D.L.; Staley, C.; Gould, T.J.; Sadowsky, M.J. Associations between soil bacterial community structure and nutrient cycling functions in long-term organic farm soils following cover crop and organic fertilizer amendment. Sci. Total Environ. 2016, 566–567, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Ushiki, N.; Fujitani, H.; Aoi, Y.; Tsuneda, S. Isolation of Nitrospira belonging to sublineage ii from a wastewater treatment plant. Microbes Environ. 2013, 28, 346–353. [Google Scholar] [CrossRef]
- Kim, S.-J.; Ahn, J.-H.; Lee, T.-H.; Weon, H.-Y.; Hong, S.-B.; Seok, S.-J.; Whang, K.-S.; Kwon, S.-W. Reyranella soli sp. Nov., isolated from forest soil, and emended description of the genus Reyranella pagnier et al. 2011. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 9, 3164–3167. [Google Scholar] [CrossRef]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; He, Y.; Liu, H.; Dumont, M.G.; Brookes, P.C.; Xu, J. Nitrosospira cluster 3-like bacterial ammonia oxidizers and nitrospira-like nitrite oxidizers dominate nitrification activity in acidic terrace paddy soils. Soil Biol. Biochem. 2019, 131, 229–237. [Google Scholar] [CrossRef]
- Mazzoncini, M.; Sapkota, T.B.; Bàrberi, P.; Antichi, D.; Risaliti, R. Long-term effect of tillage, nitrogen fertilization and cover crops on soil organic carbon and total nitrogen content. Soil Tillage Res. 2011, 114, 165–174. [Google Scholar] [CrossRef]
- Hubbard, R.K.; Strickland, T.C.; Phatak, S. Effects of cover crop systems on soil physical properties and carbon/nitrogen relationships in the coastal plain of southeastern USA. Soil Tillage Res. 2013, 126, 276–283. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Clayton, G.W.; O’Donovan, J.T.; Harker, K.N.; Turkington, T.K.; Rice, W.A. Decomposition of crop residues under conventional and zero tillage. Can. J. Soil Sci. 2004, 84, 403–410. [Google Scholar] [CrossRef]
- Lynch, M.J.; Mulvaney, M.J.; Hodges, S.C.; Thompson, T.L.; Thomason, W.E. Decomposition, nitrogen and carbon mineralization from food and cover crop residues in the central plateau of Haiti. SpringerPlus 2016, 5, 973. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, D.-U.; Lee, S.; Park, S.; Yoon, J.-H.; Seong, C.N.; Ka, J.-O. Reyranella terrae sp. Nov., isolated from an agricultural soil, and emended description of the genus Reyranella. Int. J. Syst. Evol. Microbiol. 2017, 67, 2031–2035. [Google Scholar] [CrossRef] [PubMed]
- de Vries, M.; Schöler, A.; Ertl, J.; Xu, Z.; Schloter, M. Metagenomic analyses reveal no differences in genes involved in cellulose degradation under different tillage treatments. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Villanueva, D.A.; Bello-López, J.M.; Navarro-Noya, Y.E.; Luna-Guido, M.; Verhulst, N.; Govaerts, B.; Dendooven, L. Bacterial community structure in maize residue amended soil with contrasting management practices. Appl. Soil Ecol. 2015, 90, 49–59. [Google Scholar] [CrossRef]
- Schmidt, R.; Mitchell, J.; Scow, K. Cover cropping and no-till increase diversity and symbiotroph:Saprotroph ratios of soil fungal communities. Soil Biol. Biochem. 2019, 129, 99–109. [Google Scholar] [CrossRef]
- Yadav, A.N.; Kumar, V.; Dhaliwal, H.S.; Prasad, R.; Saxena, A.K. Microbiome in Crops: Diversity, Distribution, and Potential Role in Crop Improvement; Prasad, R., Gill, S.S., Tuteja, N., Eds.; Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–332. [Google Scholar]
- De Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Kerou, M.; Schleper, C. Nitrososphaera; Bergey’s Manual Trust; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 1–10. [Google Scholar]
- Taylor, A.E.; Zeglin, L.H.; Dooley, S.; Myrold, D.D.; Bottomley, P.J. Evidence for different contributions of archaea and bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl. Environ. Microbiol. 2010, 76, 7691–7698. [Google Scholar] [CrossRef]
- Zhalnina, K.; De Quadros, P.D.; Gano, K.A.; Davis-Richardson, A.; Fagen, J.R.; Brown, C.T.; Giongo, A.; Drew, J.C.; Sayavedra-Soto, L.A.; Arp, D.J.; et al. Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front. Microbiol. 2013, 4, 104. [Google Scholar] [CrossRef]
- Perrone, S.; Grossman, J.; Liebman, A.; Sooksa-nguan, T.; Gutknecht, J. Nitrogen fixation and productivity of winter annual legume cover crops in upper Midwest organic cropping systems. Nutr. Cycl. Agroecosyst. 2020, 117, 1–16. [Google Scholar] [CrossRef]
- Chen, R.; Senbayram, M.; Blagodatsky, S.; Myachina, O.; Dittert, K.; Lin, X.; Blagodatskaya, E.; Kuzyakov, Y. Soil C and N availability determine the priming effect: Microbial N mining and stoichiometric decomposition theories. Glob. Chang. Biol. 2014, 20, 2356–2367. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.D.; Avis, P.G.; Reich, P.B.; Hobbie, S.E. Positive feedbacks between decomposition and soil nitrogen availability along fertility gradients. Plant Soil 2013, 367, 347–361. [Google Scholar] [CrossRef]
- Sainju, U.M.; Whitehead, W.F.; Singh, B.P. Biculture legume–cereal cover crops for enhanced biomass yield and carbon and nitrogen. Agron. J. 2005, 97, 1403–1412. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Friedel, J.K.; Stahr, K. Review of mechanisms and quantification of priming effects. Soil Biol. Biochem. 2000, 32, 1485–1498. [Google Scholar] [CrossRef]
- Gonzalez, J.M. Runoff and losses of nutrients and herbicides under long-term conservation practices (no-till and crop rotation) in the U.S. Midwest: A variable intensity simulated rainfall approach. Int. Soil Water Conserv. Res. 2018, 6, 265–274. [Google Scholar] [CrossRef]
- Shackelford, G.E.; Kelsey, R.; Dicks, L.V. Effects of cover crops on multiple ecosystem services: Ten meta-analyses of data from arable farmland in California and the Mediterranean. Land Use Policy 2019, 88, 104204. [Google Scholar] [CrossRef]
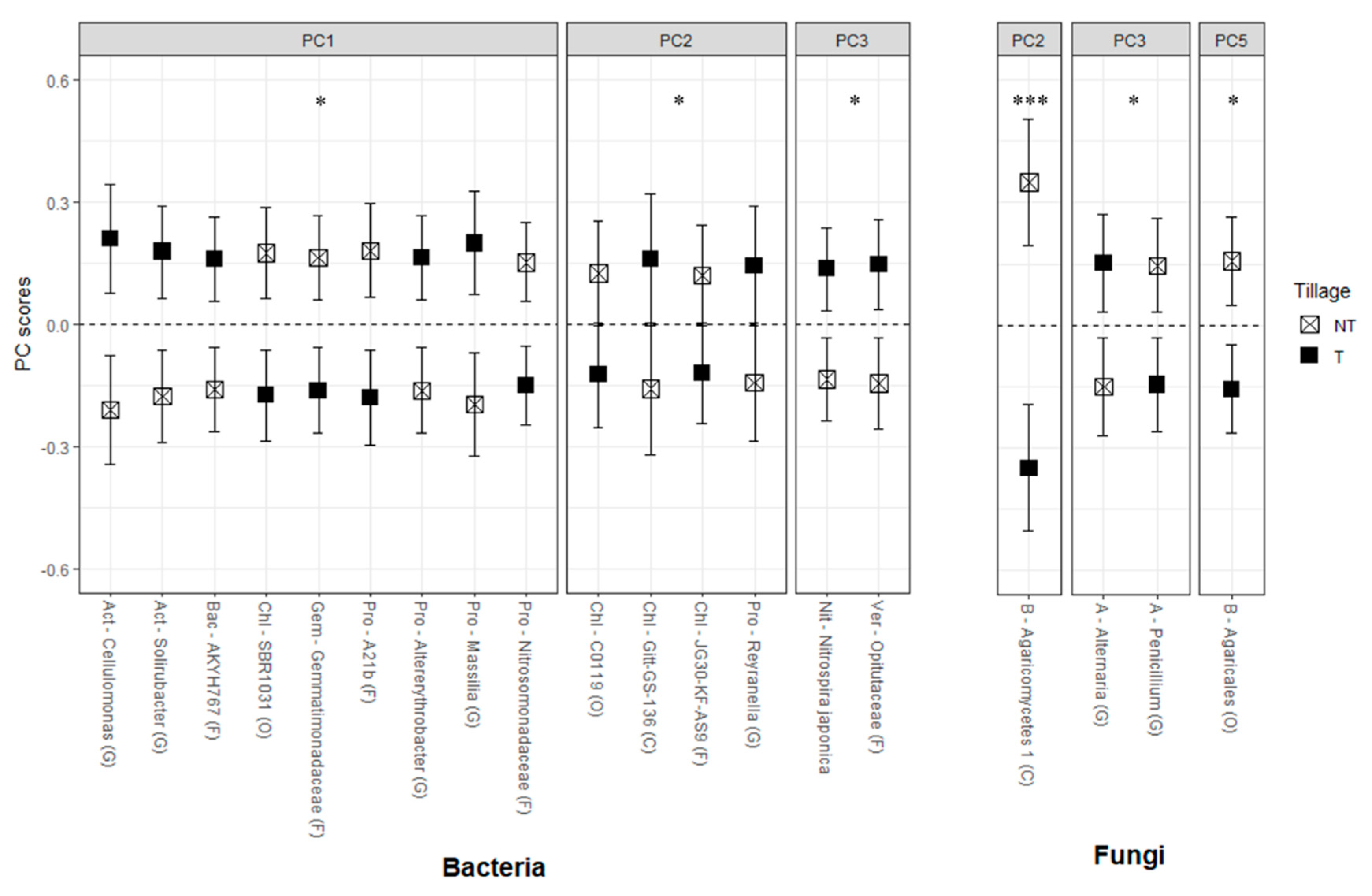
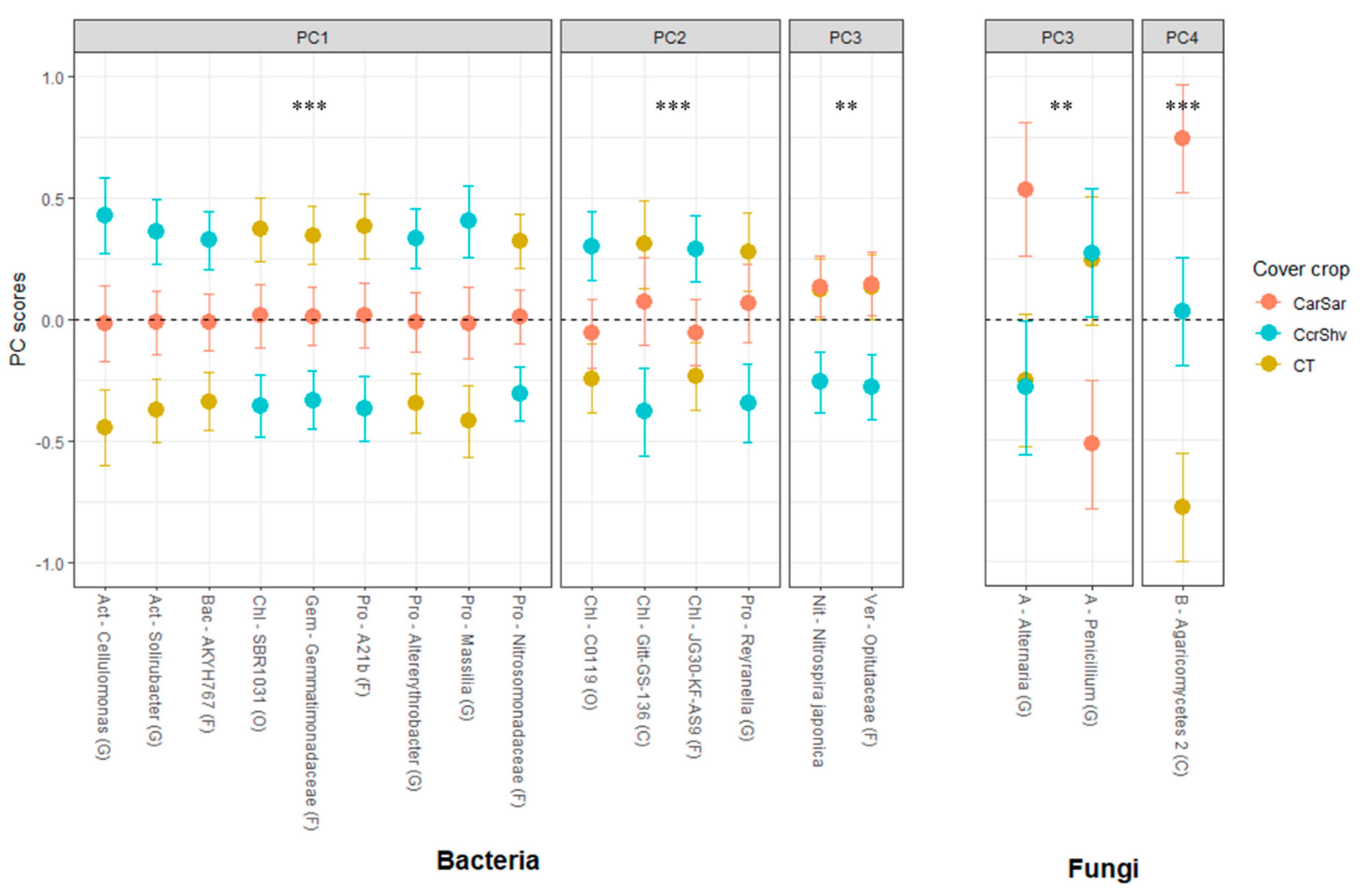
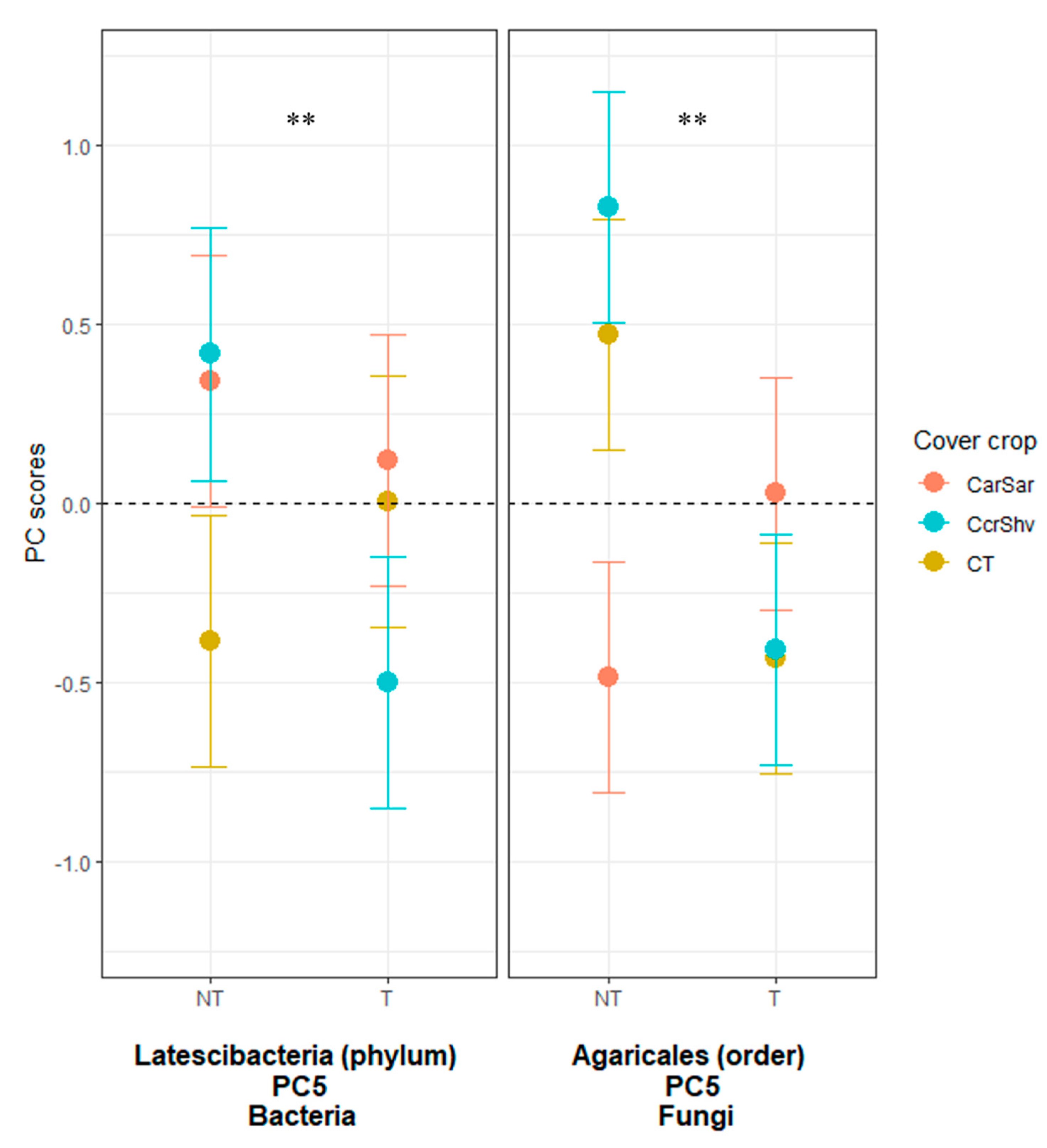
| Tillage † | Cover Crop ‡ | ||||||
|---|---|---|---|---|---|---|---|
| Soil Properties | NT | T | p-Value | CT | CarSar | CcrShv | p-Value |
| CEC | 21.78 (2.49) | 24.04 (2.49) | 0.43 | 24.07 (2.57) | 21.59 (2.57) | 23.06 (2.57) | 0.63 |
| pH | 5.95 (0.16) | 5.90 (0.16) | 0.72 | 5.96 (0.16) | 5.94 (0.16) | 5.88 (0.16) | 0.74 |
| SOM | 37.6 (1.50) | 38.3 (1.50) | 0.34 | 37.3 (1.50) | 38.1 (1.50) | 38.5 (1.50) | 0.38 |
| NO3-N | 1.52 (0.23) | 1.47 (0.23) | 0.85 | 1.83a (0.33) | 0.85b (0.33) | 1.80a (0.33) | 0.07 |
| NH4-N | 12.62 (0.81) | 11.90 (0.81) | 0.5 | 11.84 (1.06) | 12.68 (1.06) | 12.27 (1.06) | 0.85 |
| P | 5.42 (1.51) | 5.79 (1.51) | 0.74 | 6.38 (1.56) | 4.31 (1.56) | 6.13 (1.56) | 0.18 |
| Cover Crop Biomass | |||||||
| C | 427.7a (5.00) | 414.6b (4.70) | 0.04 | 413.8b (5.30) | 428.5a (5.90) | 0.08 | |
| N | 29.3 (1.60) | 26.5 (1.50) | 0.14 | 27.9 (1.60) | 27.9 (1.70) | 1.00 | |
| C:N | 15.43 (0.71) | 15.91 (0.66) | 0.58 | 15.24 (0.69) | 16.10 (0.78) | 0.41 | |
| DW | 1.80 (0.10) | 1.93 (0.09) | 0.27 | 1.81 (0.09) | 1.92 (0.10) | 0.33 | |
| OTUs | Chao1 | H’ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Taxa | Treatment † | Mean | SEM | p-Value | Mean | SEM | p-Value | Mean | SEM | p-Value |
| Bacteria | T | 206.04 | 10.14 | 0.54 | 206.35 | 10.19 | 0.54 | 7.3 | 0.07 | 0.57 |
| NT | 210.11 | 210.47 | 7.33 | |||||||
| CarSar | 202.46 | 10.62 | 0.38 | 202.72 | 10.68 | 0.37 | 7.27 | 0.08 | 0.27 | |
| CcrShv | 213.62 | 214.2 | 7.37 | |||||||
| CT | 208.15 | 208.31 | 7.32 | |||||||
| Fungi | T | 32.94 | 1.52 | 0.26 | 32.97 | 1.55 | 0.28 | 4.19 | 0.08 | 0.11 |
| NT | 30.37 | 30.45 | 4.01 | |||||||
| CarSar | 31.95 | 1.56 | 0.89 | 31.97 | 1.58 | 0.9 | 4.26 | 0.12 | 0.3 | |
| CcrShv | 31.9 | 31.96 | 4.02 | |||||||
| CT | 31.11 | 31.2 | 4.03 | |||||||
| Archaea | T | 11.6 | 0.65 | 0.58 | 11.6 | 0.65 | 0.58 | 3.24 | 0.08 | 0.73 |
| NT | 11.16 | 11.16 | 3.2 | |||||||
| CarSar | 11.45 | 0.72 | 0.98 | 11.45 | 0.73 | 0.97 | 3.24 | 0.09 | 0.76 | |
| CcrShv | 11.41 | 11.43 | 3.24 | |||||||
| CT | 11.29 | 11.26 | 3.17 | |||||||
| Taxa | Treatment | Levels Compared † | Sample Size | Pseudo-F | p-Value | q-Value |
|---|---|---|---|---|---|---|
| Bacteria | Till | NT–T | 142 | 4.33 | 0.001 | 0.001 |
| CC | CT–CarSar | 95 | 1.08 | 0.296 | 0.444 | |
| CT–CcrShv | 94 | 1.30 | 0.192 | 0.444 | ||
| CarSar–CcrShv | 95 | 0.77 | 0.774 | 0.774 | ||
| Fungi | Till | NT–T | 129 | 2.78 | 0.019 | 0.019 |
| CC | CT–CarSar | 83 | 5.42 | 0.002 | 0.003 | |
| CT–CcrShv | 87 | 3.14 | 0.013 | 0.013 | ||
| CarSar–CcrShv | 88 | 4.11 | 0.001 | 0.003 | ||
| Archaea | Till | NT–T | 130 | 1.12 | 0.272 | 0.272 |
| CC | CT–CarSar | 86 | 2.90 | 0.054 | 0.081 | |
| CT–CcrShv | 87 | 3.98 | 0.01 | 0.03 | ||
| CarSar–CcrShv | 87 | 0.80 | 0.437 | 0.437 |
| Bacteria | Fungi | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | ||
| Treatments | df | p-Value | p-Value | |||||||||||
| Till | 1 | 0.062 | 0.082 | 0.075 | 0.850 | 0.465 | 0.815 | 0.213 | 0.252 | 0.005 | 0.059 | 0.145 | 0.080 | 0.964 |
| CC | 2 | 0.004 | 0.009 | 0.045 | 0.231 | 0.485 | 0.128 | 0.173 | 0.873 | 0.517 | 0.044 | 0.001 | 0.419 | 0.796 |
| CC × Till | 2 | 0.968 | 0.101 | 0.131 | 0.363 | 0.014 | 0.025 | 0.285 | 0.682 | 0.339 | 0.128 | 0.497 | 0.039 | 0.171 |
| Treatment means † | ||||||||||||||
| NT | −0.29 | −0.22 | −0.26 | 0.03 | 0.12 | −0.03 | −0.18 | −0.16 | −0.47b | −0.27 | 0.20 | 0.27 | −0.01 | |
| T | 0.29 | 0.22 | 0.26 | −0.03 | −0.12 | 0.03 | 0.18 | 0.16 | 0.47a | 0.27 | −0.20 | −0.27 | 0.01 | |
| SEM | 0.19 | 0.23 | 0.20 | 0.25 | 0.27 | 0.19 | 0.22 | 0.25 | 0.21 | 0.21 | 0.17 | 0.19 | 0.21 | |
| CT | −0.62b | 0.43a | 0.24a | 0.24 | −0.19 | 0.44 | −0.19 | 0.09 | −0.08 | −0.23b | −0.72c | 0.02 | 0.06 | |
| CarSar | −0.02a | 0.10a | 0.26a | −0.31 | 0.23 | −0.16 | 0.38 | 0.00 | −0.12 | 0.48a | 0.69a | −0.23 | −0.14 | |
| CcrShv | 0.59a | −0.54b | −0.50b | 0.07 | −0.04 | −0.27 | −0.19 | −0.09 | 0.21 | −0.25b | 0.03b | 0.21 | 0.08 | |
| SEM | 0.22 | 0.25 | 0.24 | 0.25 | 0.29 | 0.25 | 0.26 | 0.29 | 0.24 | 0.24 | 0.21 | 0.23 | 0.25 | |
| NT × CT | −0.87 | 0.44 | −0.34 | 0.03 | −0.39ab | 0.24ab | −0.07 | −0.23 | −0.46 | −0.70 | −0.70 | 0.47ab | 0.00 | |
| NT × CarSar | −0.27 | −0.48 | −0.02 | −0.06 | 0.34ab | 0.33a | 0.13 | −0.11 | −0.84 | 0.04 | 1.06 | −0.49b | 0.23 | |
| NT × CcrShv | 0.27 | −0.64 | −0.42 | 0.13 | 0.42a | −0.66b | −0.60 | −0.14 | −0.10 | −0.15 | 0.23 | 0.83a | −0.25 | |
| T × CT | −0.36 | 0.42 | 0.82 | 0.46 | 0.01ab | 0.64a | −0.30 | 0.41 | 0.29 | 0.25 | −0.73 | −0.43b | 0.13 | |
| T × CarSar | 0.32 | 0.68 | 0.54 | −0.56 | 0.12ab | −0.66b | 0.62 | 0.10 | 0.60 | 0.91 | 0.32 | 0.03ab | −0.51 | |
| T × CcrShv | 0.92 | −0.43 | −0.58 | 0.00 | −0.50b | 0.11ab | 0.22 | −0.03 | 0.52 | −0.35 | −0.17 | −0.41b | 0.41 | |
| SEM | 0.30 | 0.32 | 0.32 | 0.36 | 0.35 | 0.32 | 0.34 | 0.37 | 0.32 | 0.32 | 0.29 | 0.32 | 0.36 | |
| Archaea | ||||||
|---|---|---|---|---|---|---|
| C. Nitrososphaera | SCA1154 | SCA1158 | SCA1166 | SCA1173 | ||
| Factors | df | p-Value | ||||
| Till | 1 | 0.533 | 0.236 | 0.181 | 0.107 | 0.248 |
| CC | 2 | 0.129 | 0.174 | 0.588 | 0.533 | 0.023 |
| CC × Till | 2 | 0.428 | 0.185 | 0.211 | 0.698 | 0.478 |
| Treatment means † | ||||||
| NT | 0.09 | 0.11 | 0.06 | 0.37 | 0.21 | |
| T | 0.13 | 0.06 | 0.12 | 0.24 | 0.30 | |
| SEM | 0.04 | 0.04 | 0.04 | 0.06 | 0.06 | |
| CT | 0.03 | 0.04 | 0.12 | 0.30 | 0.40a | |
| CarSar | 0.17 | 0.05 | 0.07 | 0.37 | 0.13b | |
| CcrShv | 0.12 | 0.16 | 0.07 | 0.25 | 0.24ab | |
| SEM | 0.05 | 0.05 | 0.04 | 0.06 | 0.07 | |
| NT × CT | 0.04 | 0.03 | 0.03 | 0.39 | 0.40 | |
| NT × CarSar | 0.18 | 0.07 | 0.05 | 0.39 | 0.09 | |
| NT × CcrShv | 0.06 | 0.23 | 0.08 | 0.32 | 0.15 | |
| T × CT | 0.03 | 0.06 | 0.20 | 0.21 | 0.39 | |
| T × CarSar | 0.17 | 0.04 | 0.09 | 0.35 | 0.18 | |
| T × CcrShv | 0.19 | 0.09 | 0.06 | 0.18 | 0.33 | |
| SEM | 0.06 | 0.06 | 0.05 | 0.09 | 0.09 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Zabaloy, M.C.; Riggins, C.W.; Rodríguez-Zas, S.; Villamil, M.B. Microbial Shifts Following Five Years of Cover Cropping and Tillage Practices in Fertile Agroecosystems. Microorganisms 2020, 8, 1773. https://doi.org/10.3390/microorganisms8111773
Kim N, Zabaloy MC, Riggins CW, Rodríguez-Zas S, Villamil MB. Microbial Shifts Following Five Years of Cover Cropping and Tillage Practices in Fertile Agroecosystems. Microorganisms. 2020; 8(11):1773. https://doi.org/10.3390/microorganisms8111773
Chicago/Turabian StyleKim, Nakian, María C. Zabaloy, Chance W. Riggins, Sandra Rodríguez-Zas, and María B. Villamil. 2020. "Microbial Shifts Following Five Years of Cover Cropping and Tillage Practices in Fertile Agroecosystems" Microorganisms 8, no. 11: 1773. https://doi.org/10.3390/microorganisms8111773
APA StyleKim, N., Zabaloy, M. C., Riggins, C. W., Rodríguez-Zas, S., & Villamil, M. B. (2020). Microbial Shifts Following Five Years of Cover Cropping and Tillage Practices in Fertile Agroecosystems. Microorganisms, 8(11), 1773. https://doi.org/10.3390/microorganisms8111773







