Comparison of DNA Extraction Methods and Real-Time PCR Assays for the Detection of Blastocystis sp. in Stool Specimens
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. DNA Extraction
2.3. Comparison of the Four qPCR Assays
2.4. Blastocystis Subtype Sequencing
2.5. Statistical Analysis
3. Results
3.1. Specimens
3.2. Comparison of DNA Extraction Procedures
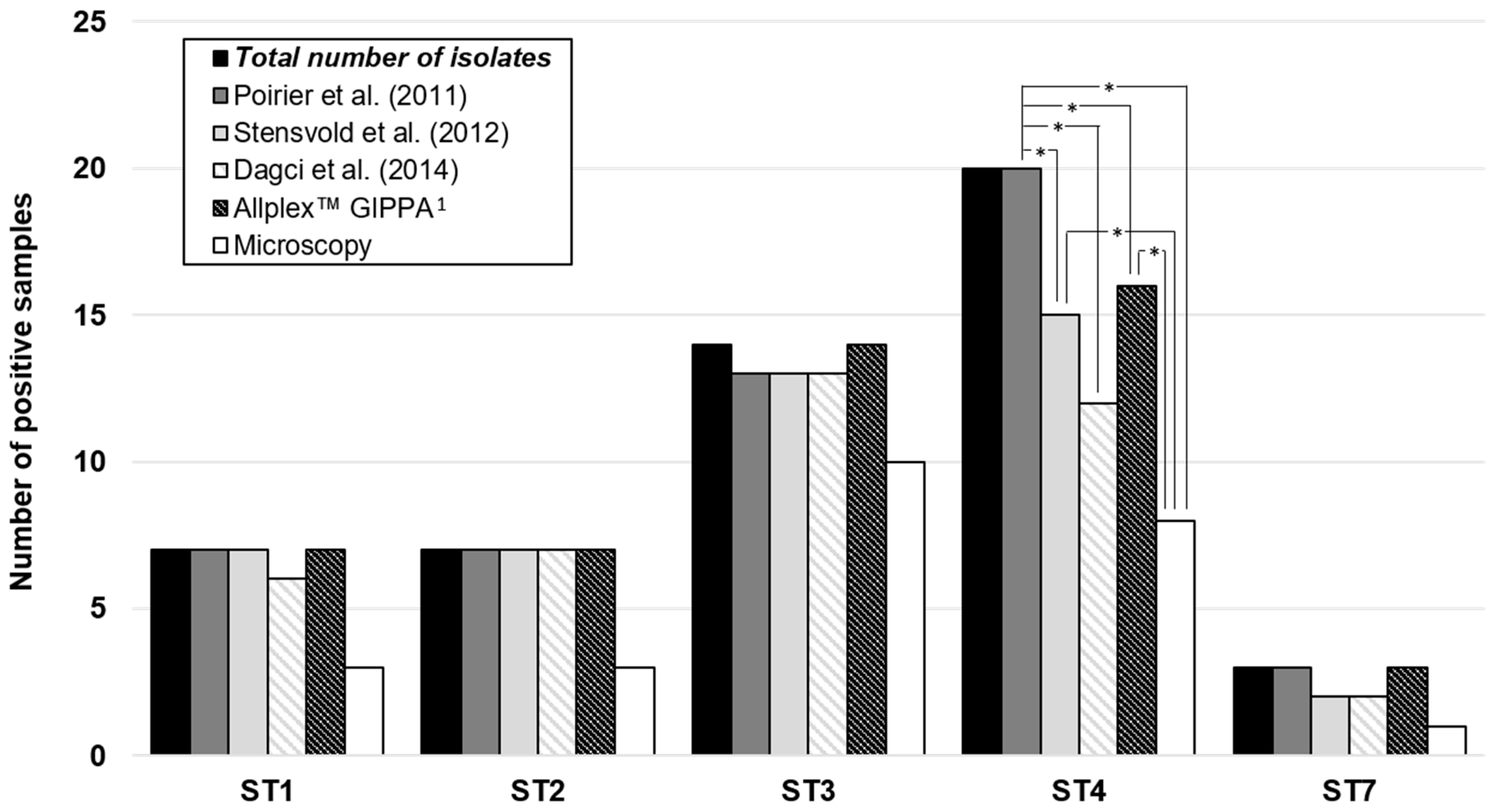
3.3. Comparison of the Four qPCR Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stensvold, C.R.; Clark, C.G. Pre-empting Pandora’s box: Blastocystis subtypes revisited. Trends Parasitol. 2020, 36, 229–232. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Taner-Mulla, D.; Jacob, A.S.; Imeede, C.A.; Yoshikawa, H.; Stensvold, C.R.; Graham, C. Genetic Diversity of Blastocystis in Livestock and Zoo Animals. Protist 2013, 164, 497–509. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Sánchez, A.; Hernández, C.; Flórez, C.; Bernal, M.C.; Giraldo, J.C.; Reyes, P.; Lopez, M.; Garcia, L.; Cooper, P.; et al. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016, 41, 32–35. [Google Scholar] [CrossRef]
- Tan, K.S.W. New Insights on Classification, Identification, and Clinical Relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665. [Google Scholar] [CrossRef]
- Nourrisson, C.; Scanzi, J.; Pereira, B.; NkoudMongo, C.; Wawrzyniak, I.; Cian, A.; Viscogliosi, E.; Livrelli, V.; Delbac, F.; Dapoigny, M.; et al. Blastocystis is associated with decrease of fecal microbiota protective bacteria: Comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE 2014, 9, e111868. [Google Scholar] [CrossRef]
- Nagel, R.; Traub, R.J.; Allcock, R.J.N.; Kwan, M.M.S.; Bielefeldt-Ohmann, H. Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome 2016, 4, 47. [Google Scholar] [CrossRef]
- Forsell, J.; Bengtsson-Palme, J.; Angelin, M.; Johansson, A.; Evengård, B.; Granlund, M. The relation between Blastocystis and the intestinal microbiota in Swedish travellers. BMC Microbiol. 2017, 17, 231. [Google Scholar] [CrossRef]
- Nieves-Ramírez, M.E.; Partida-Rodríguez, O.; Laforest-Lapointe, I.; Reynolds, L.A.; Brown, E.M.; Valdez-Salazar, A.; Moan-Silva, P.; Rojas-Velazquez, L.; Morien, E.; Parfrey, L.W.; et al. Asymptomatic Intestinal Colonization with Protist Blastocystis Is Strongly Associated with Distinct Microbiome Ecological Patterns. mSystems 2018, 3. [Google Scholar] [CrossRef]
- Audebert, C.; Even, G.; Cian, A.; Blastocystis Investigation Group; Loywick, A.; Merlin, S.; Viscogliosi, E.; Chabé, M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 2016, 6, 25255. [Google Scholar] [CrossRef]
- Defaye, M.; Nourrisson, C.; Baudu, E.; Warwzyniak, I.; Bonnin, V.; Bonnet, M.; Barnich, N.; Ardid, D.; Delbac, F.; Carvalho, F.A.; et al. Efficient and reproducible experimental infections of rats with Blastocystis spp. PLoS ONE 2018, 13, e0207669. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilić-Stojanović, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Terveer, E.M.; van Gool, T.; Ooijevaar, R.E.; Sanders, I.M.J.G.; Boeije-Koppenol, E.; Keller, J.J.; Bart, A.; Kuijper, E.; Terveer, E.; Vendrik, K.; et al. Human transmission of Blastocystis by Fecal Microbiota Transplantation without development of gastrointestinal symptoms in recipients. Clin. Infect. Dis. 2019, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Poirier, P.; Wawrzyniak, I.; Albert, A.; El Alaoui, H.; Delbac, F.; Livrelli, V. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: Prospective study of patients with hematological malignancies. J. Clin. Microbiol. 2011, 49, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.S., 2nd; Ganac, R.D.; Hiser, G.; Hudson, N.R.; Le, A.; Whipps, C.M. Detection of Blastocystis from stool samples using real-time PCR. Parasitol. Res. 2008, 103, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Ahmed, U.N.; Andersen, L.O.; Nielsen, H.V. Development and evaluation of a genus-specific, probe-based, internal-process-controlled real-time PCR assay for sensitive and specific detection of Blastocystis spp. J. Clin. Microbiol. 2012, 50, 1847–1851. [Google Scholar] [CrossRef]
- Dagci, H.; Kurt, Ö.; Demirel, M.; Mandiracioglu, A.; Aydemir, S.; Saz, U.; Bart, A.; van Gool, T. Epidemiological and diagnostic features of Blastocystis infection in symptomatic patients in izmir province, Turkey. Iran J. Parasitol. 2014, 9, 519–529. [Google Scholar]
- El Safadi, D.; Cian, A.; Nourrisson, C.; Pereira, B.; Morelle, C.; Bastien, P.; Bellanger, A.P.; Botterel, F.; Candolfi, E.; Desoubeaux, G.; et al. Prevalence, risk factors for infection and subtype distribution of the intestinal parasite Blastocystis sp. from a large-scale multi-center study in France. BMC Infect. Dis. 2016, 16, 451. [Google Scholar] [CrossRef]
- Laude, A.; Valot, S.; Desoubeaux, G.; Argy, N.; Nourrisson, C.; Pomares, C.; Machouart, M.; Le Govic, Y.; Dalle, F.; Botterel, F.; et al. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin. Microbiol. Infect. 2016, 22, 190.e1–190.e8. [Google Scholar]
- Van Zanten, E.; Wisselink, G.J.; de Boer, W.; Stoll, S.; Alvarez, R.; Kooistra-Smid, A.M.D. Comparison of the QIAsymphony automated nucleic acid extraction and PCR setup platforms with NucliSens easyMAG and Corbett CAS-1200 liquid handling station for the detection of enteric pathogens in fecal samples. J. Microbiol. Methods 2011, 84, 335–340. [Google Scholar] [CrossRef]
- Roux, G.; Varlet-Marie, E.; Bastien, P.; Sterkers, Y. French National Reference Center for Toxoplasmosis Network. Evolution of Toxoplasma-PCR methods and practices: A French national survey and proposal for technical guidelines. Int. J. Parasitol. 2018, 48, 701–707. [Google Scholar] [CrossRef]
- Scicluna, S.M.; Tawari, B.; Clark, C.G. DNA barcoding of Blastocystis. Protist 2006, 157, 77–85. [Google Scholar] [CrossRef] [PubMed]

| Poirier et al. [12] | Stensvold et al. [14] | Dagci et al. [15] | AllplexTM GIPPA 1 | Direct Light Microscopy | |
|---|---|---|---|---|---|
| Sensitivity | 0.71 | 0.79 | 0.55 | 0.84 | 0.34 |
| Specificity | 0.98 | 0.94 | 1.00 | 0.82 | 1.00 |
| Poirier et al. [12] | Stensvold et al. [14] | Dagci et al. [15] | AllplexTM GIPPA 1 | |
|---|---|---|---|---|
| Probe-based qPCR | n.a. 2 | Taq-Man probe | Taq-Man probe | TOCETM technology 3 |
| Internal control | n.a. 2 | yes | not designed in the article | yes |
| CE-IVD marking 4 | n.a. 2 | n.a. 2 | n.a. 2 | yes |
| Subtyping on amplicons | yes | n.a. 2 | n.a. 2 | n.a. 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nourrisson, C.; Brunet, J.; Flori, P.; Moniot, M.; Bonnin, V.; Delbac, F.; Poirier, P. Comparison of DNA Extraction Methods and Real-Time PCR Assays for the Detection of Blastocystis sp. in Stool Specimens. Microorganisms 2020, 8, 1768. https://doi.org/10.3390/microorganisms8111768
Nourrisson C, Brunet J, Flori P, Moniot M, Bonnin V, Delbac F, Poirier P. Comparison of DNA Extraction Methods and Real-Time PCR Assays for the Detection of Blastocystis sp. in Stool Specimens. Microorganisms. 2020; 8(11):1768. https://doi.org/10.3390/microorganisms8111768
Chicago/Turabian StyleNourrisson, Céline, Julie Brunet, Pierre Flori, Maxime Moniot, Virginie Bonnin, Frédéric Delbac, and Philippe Poirier. 2020. "Comparison of DNA Extraction Methods and Real-Time PCR Assays for the Detection of Blastocystis sp. in Stool Specimens" Microorganisms 8, no. 11: 1768. https://doi.org/10.3390/microorganisms8111768
APA StyleNourrisson, C., Brunet, J., Flori, P., Moniot, M., Bonnin, V., Delbac, F., & Poirier, P. (2020). Comparison of DNA Extraction Methods and Real-Time PCR Assays for the Detection of Blastocystis sp. in Stool Specimens. Microorganisms, 8(11), 1768. https://doi.org/10.3390/microorganisms8111768





