In Vivo and In Vitro Virulence Analysis of Four Genetically Distinct Toxoplasma gondii Lineage III Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Genotyping and Virulence Marker Typing by MnPCR-RFLP
2.3. Mice
2.4. In Vivo Conversion of Bradyzoites to Tachyzoites and Tachyzoite Propagation in Cell Culture
2.5. In Vivo Survival Assay
2.6. In Vitro Growth and Plaque Assay
2.7. RNA Extraction, Reverse Transcription, and Relative Quantification
2.8. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dubey, J.P.; Lindsay, D.S.; Speer, C.A. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 1998, 11, 267–299. [Google Scholar] [CrossRef] [PubMed]
- Halonen, S.K.; Weiss, L.M. Toxoplasmosis. Handb. Clin. Neurol. 2013, 114, 125–145. [Google Scholar] [CrossRef]
- Mendez, O.A.; Koshy, A.A. Toxoplasma gondii: Entry, association, and physiological influence on the central nervous system. PLoS Pathog. 2017, 13, e1006351. [Google Scholar] [CrossRef] [PubMed]
- Shwab, E.; Zhu, X.; Majumdar, D.; Pena, H.; Gennari, S.; Dubey, J.; Su, C. Geographical patterns of Toxoplasma gondii genetic diversity revealed by multilocus PCR-RFLP genotyping. Parasitology 2014, 141, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Dumètre, A.; Dubey, J.P.; Ferguson, D.J.P.; Bongrand, P.; Azas, N.; Puech, P.-H. Mechanics of the Toxoplasma gondii oocyst wall. Proc. Natl. Acad. Sci. USA 2013, 110, 11535–11540. [Google Scholar] [CrossRef]
- Hill, D.; Dubey, J.P. Toxoplasma gondii: Transmission, diagnosis and prevention. Clin. Microbiol. Inf. 2002, 8, 634–640. [Google Scholar] [CrossRef]
- Aguirre, A.A.; Longcore, T.; Barbieri, M.; Dabritz, H.; Hill, D.; Klein, P.N.; Lepczyk, C.; Lilly, E.L.; McLeod, R.; Milcarsky, J.; et al. The One Health Approach to Toxoplasmosis: Epidemiology, Control, and Prevention Strategies. EcoHealth 2019, 16, 378–390. [Google Scholar] [CrossRef]
- Fuglewicz, A.J.; Piotrowski, P.; Stodolak, A. Relationship between toxoplasmosis and schizophrenia: A review. Adv. Clin. Exp. Med. 2017, 26, 1031–1036. [Google Scholar] [CrossRef]
- Robert-Gangneux, F.; Meroni, V.; Dupont, D.; Botterel, F.; Garcia, J.M.A.; Brenier-Pinchart, M.P.; Accoceberry, I.; Akan, H.; Abbate, I.; Boggian, K.; et al. Toxoplasmosis in Transplant Recipients, Europe, 2010–2014. Emerg. Infect. Dis. 2018, 24, 1497–1504. [Google Scholar] [CrossRef]
- Howe, D.K.; Sibley, L.D. Toxoplasma gondii comprises three clonal lineages: Correlation of parasite genotype with human disease. J. Infect. Dis. 1995, 172, 1561–1566. [Google Scholar] [CrossRef]
- Dardé, M.L. Toxoplasma gondii, “New” Genotypes and Virulence. Parasite 2008, 15, 366–371. [Google Scholar] [CrossRef]
- Galal, L.; Hamidovic, A.; Dardé, M.L.; Mercier, A. Diversity of Toxoplasma gondii strains at the global level and its determinants. Food Waterborne Parasitol. 2019, 15, e00052. [Google Scholar] [CrossRef] [PubMed]
- Peyron, F.; Lobry, J.R.; Musset, K.; Ferrandiz, J.; Gomez-Marin, J.E.; Petersen, E.; Meroni, V.; Rausher, B.; Mercier, C.; Picot, S.; et al. Serotyping of Toxoplasma gondii in chronically infected pregnant women: Predominance of type II in Europe and types I and III in Colombia (South America). Microbes Infect. 2006, 8, 2333–2340. [Google Scholar] [CrossRef] [PubMed]
- Vilares, A.; Gargaté, M.J.; Ferreira, I.; Martins, S.; Gomes, J.P. Molecular and virulence characterization of Toxoplasma gondii strains isolated from humans in Portugal. Parasitol Res. 2017, 116, 979–985. [Google Scholar] [CrossRef]
- Simon, S.; de Thoisy, B.; Mercier, A.; Nacher, M.; Demar, M. Virulence of atypical Toxoplasma gondii strains isolated in French Guiana in a murine model. Parasite 2019, 26, 60. [Google Scholar] [CrossRef]
- Djurković-Djaković, O.; Klun, I.; Khan, A.; Nikolić, A.; Knezević-Ušaj, S.; Bobić, B.; Sibley, L.D. A human origin type II strain of Toxoplasma gondii causing severe encephalitis in mice. Microbes Infect. 2006, 8, 2206–2212. [Google Scholar] [CrossRef] [PubMed]
- Štajner, T.; Vasiljević, Z.; Vujić, D.; Marković, M.; Ristić, G.; Mićić, D.; Pašić, S.; Ivović, V.; Ajzenberg, D.; Djurković-Djaković, O. Atypical strain of Toxoplasma gondii causing fatal reactivation after hematopoietic stem cell transplantion in a patient with an underlying immunological deficiency. J. Clin. Microbiol. 2013, 51, 2686–2690. [Google Scholar] [CrossRef]
- Marković, M.; Ivović, V.; Štajner, T.; Djokić, V.; Klun, I.; Bobić, B.; Nikolić, A.; Djurković-Djaković, O. Evidence for genetic diversity of Toxoplasma gondii in selected intermediate hosts in Serbia. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 173–179. [Google Scholar] [CrossRef]
- Klun, I.; Uzelac, A.; Villena, I.; Mercier, A.; Bobić, B.; Nikolić, A.; Rajnpreht, I.; Opsteegh, M.; Aubert, D.; Blaga, R.; et al. The first isolation and molecular characterization of Toxoplasma gondii from horses in Serbia. Parasit. Vectors 2017, 10, 167. [Google Scholar] [CrossRef]
- Kuruca, L.; Uzelac, A.; Klun, I.; Lalošević, V.; Djurković-Djaković, O. Toxoplasma gondii genotypes circulating in domestic pigs in Serbia. Acta Vet. Hung. 2019, 67, 204–211. [Google Scholar] [CrossRef]
- Uzelac, A.; Klun, I.; Ćirović, D.; Penezić, A.; Ćirković, V.; Djurković-Djaković, O. Detection and genotyping of Toxoplasma gondii in wild canids in Serbia. Parasitol. Int. 2019, 73, 101973. [Google Scholar] [CrossRef] [PubMed]
- Uzelac, A.; Klun, I.; Štajner, T.; Mercier, A.; Kuruca, L.; Bobić, B.; Marković, M.; Djurković-Djaković, O. Population structure of Toxoplasma gondii in Serbia. In Proceedings of the 14th International Congress of Parasitology—ICOPA 2018, Daegu, Korea, 19–24 August 2018; pp. 3204–3206. [Google Scholar]
- Saraf, P.; Shwab, E.K.; Dubey, J.P.; Su, C. On the determination of Toxoplasma gondii virulence in mice. Exp. Parasitol. 2017, 174, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; VanWhy, K.; Verma, S.K.; Choudhary, S.; Kwok, O.C.; Khan, A.; Behnke, M.S.; Sibley, L.D.; Ferreira, L.R.; Oliveira, S.; et al. Genotyping Toxoplasma gondii from wildlife in Pennsylvania and identification of natural recombinants virulent to mice. Vet. Parasitol. 2014, 200, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Fox, B.A.; Bzik, D.J. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 2002, 415, 926–929. [Google Scholar] [CrossRef]
- Dzierszinski, F.; Mortuaire, M.; Dendouga, N.; Popescu, O.; Tomavo, S. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J. Mol. BIol. 2001, 309, 1017–1027. [Google Scholar] [CrossRef]
- Su, C.; Shwab, E.K.; Zhou, P.; Zhu, X.Q.; Dubey, J.P. Moving towards and integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010, 137, 1–11. [Google Scholar] [CrossRef]
- Desmonts, G.; Remington, J.S. Direct agglutination test for diagnosis of Toxoplasma infection: Method for increasing sensitivity and specificity. J. Clin. Microbiol. 1980, 11, 562–568. [Google Scholar] [CrossRef]
- Lélu, M.; Villena, I.; Dardé, M.L.; Aubert, D.; Geers, R.; Dupuis, E.; Marnef, F.; Poulle, M.L.; Gotteland, C.; Dumètre, A.; et al. Quantitative estimation of the viability of Toxoplasma gondii oocysts in soil. Appl. Environ. Microbiol. 2012, 78, 5127–5132. [Google Scholar] [CrossRef]
- Khan, A.; Taylor, S.; Ajioka, J.W.; Rosenthal, B.M.; Sibley, D.L. Selection at a Single Locus Leads to Widespread Expansion of Toxoplasma gondii Lineages That Are Virulent In Mice. PLoS Genet. 2009, 5, e1000404. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Quantification Strategies in real-time PCR. In A-Z of Quantitative PCR, 1st ed.; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar]
- Su, C.; Howe, D.K.; Dubey, J.P.; Ajioka, J.W.; Sibley, L.D. Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 2002, 99, 10753–10758. [Google Scholar] [CrossRef]
- Mercier, A.; Devillard, S.; Ngoubangoye, B.; Bonnabau, H.; Bañuls, A.-L.; Durand, P.; Salle, B.; Ajzenberg, D.; Dardé, M.L. Additional Haplogroups of Toxoplasma gondii out of Africa: Population Structure and Mouse-Virulence of Strains from Gabon. PLoS Negl. Trop. Dis. 2010, 4, e876. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, S.; Ajzenberg, D.; Canada, N.; Freire, L.; da Costa, J.M.C.; Dardé, M.L.; Thulliez, P.; Dubey, J.P. Biologic and molecular characterization of Toxoplasma gondii isolates from pigs from Portugal. Vet. Parasitol. 2005, 135, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Halos, L.; Thébault, A.; Aubert, D.; Thomas, M.; Perreta, C.; Geers, R.; Alliota, A.; Escotte-Binet, S.; Ajzenberg, D.; Dardé, M.L.; et al. An innovative survey underlining the significant level of contamination by Toxoplasma gondii of ovine meat consumed in France. Int. J. Parasitol. 2010, 40, 193–200. [Google Scholar] [CrossRef]
- Hamilton, C.M.; Black, L.; Oliveira, S.; Burrells, A.; Bartley, P.M.; Melo, R.; Chianini, F.; Palarea-Albaladejo, J.; Innes, E.A.; Kelly, P.J.; et al. Comparative virulence of Caribbean, Brazilian and European isolates of Toxoplasma gondii. Parasit. Vectors 2019, 12, 104. [Google Scholar] [CrossRef]
- Shwab, E.K.; Jiang, T.; Pena, H.F.; Gennari, S.M.; Dubey, J.P.; Su, C. The ROP18 and ROP5 gene allele types are highly predictive of virulence in mice across globally distributed strains of Toxoplasma gondii. Int. J. Parasitol. 2016, 46, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Saeij, J.P.J.; Boyle, J.P.; Boothroyd, J.C. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 2005, 21, 476–481. [Google Scholar] [CrossRef]
- Sullivan, W.J., Jr.; Jeffers, V. Mechanisms of Toxoplasma gondii persistence and latency. FEMS Microbiol. Rev. 2012, 36, 717–733. [Google Scholar] [CrossRef]
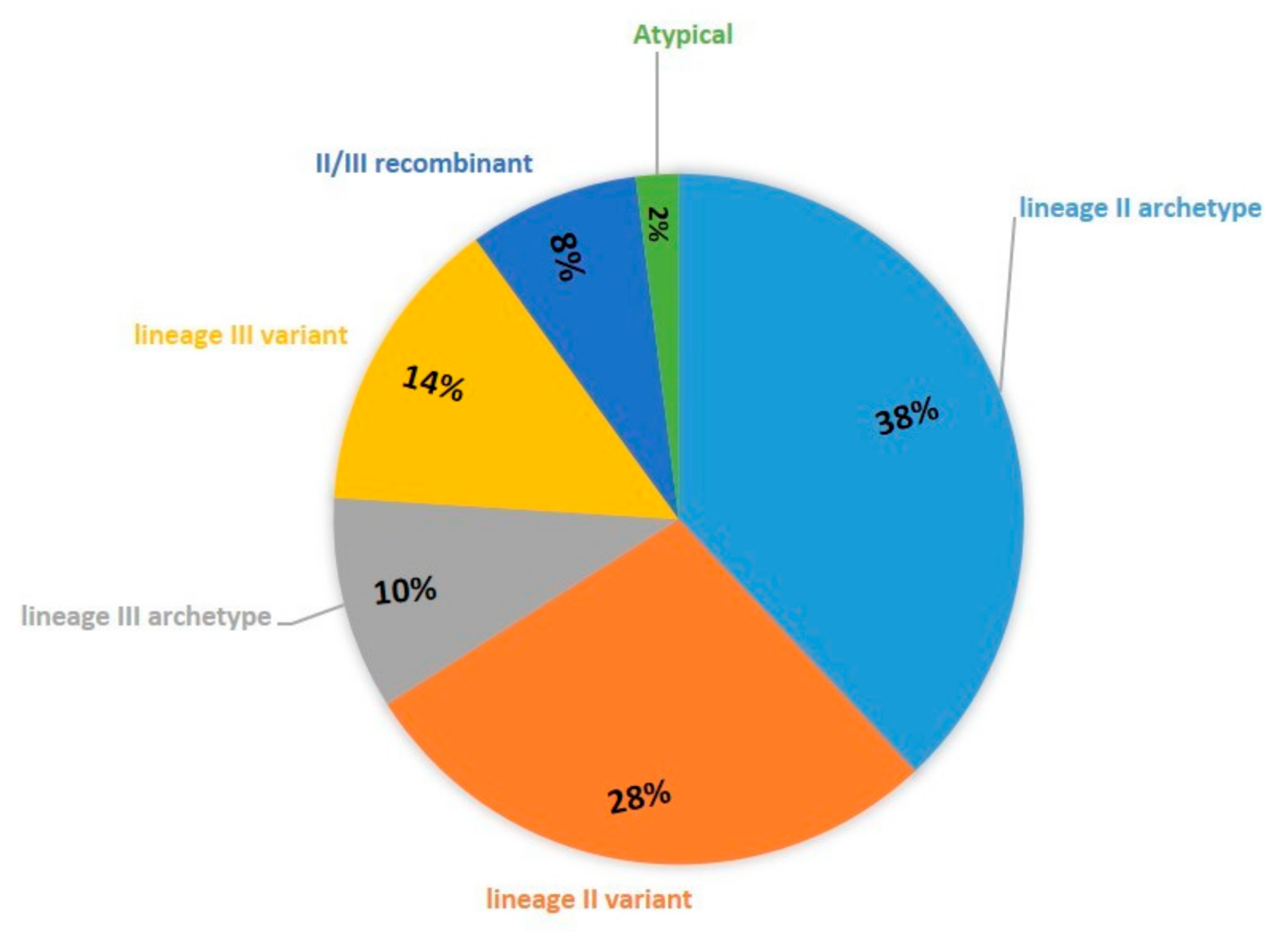


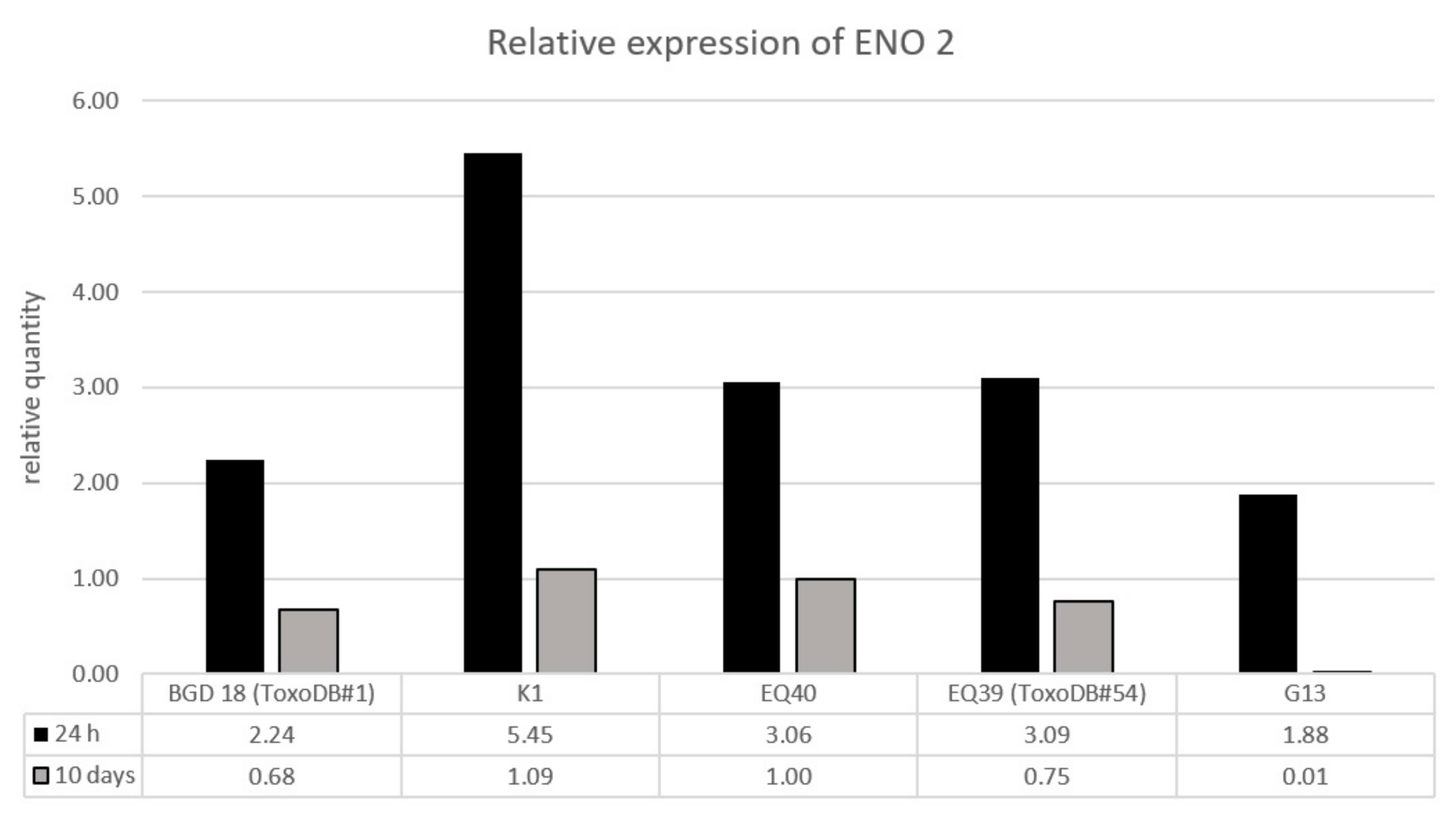
| Isolate ID | Alt. SAG2 (Chr VIII) | BTUB (Chr IX) | GRA6 (Chr X) | C22-8 (Chr Ia) | C29-2 (Chr III) | L358 (Chr V) | PK1 (Chr IV) | CS3 (Chr VIIa) | APICO (Plastid) |
|---|---|---|---|---|---|---|---|---|---|
| EQ40 | II | I | III | II | III | III | II | III | III |
| K1 | II | II | I | II | III | III | III | III | III |
| EQ39 (ToxoDB#54) | II | III | III | III | III | III | III | II | II |
| G13 | III | III | III | II | III | III | II | III | III |
| Isolate ID | Cumulative Mortality (%) at Day 42 | Virulence Classification [32] |
|---|---|---|
| EQ40 | 69.4% | Intermediately virulent |
| K1 | 38.8% | Intermediately virulent |
| EQ39 (ToxoDB#54) | 6.8% | Non-virulent |
| G13 | 10.7% | Non-virulent |
| Isolate ID | ROP5 | ROP16 | ROP18 | GRA15 |
|---|---|---|---|---|
| EQ40 | 3 | 1 | 3 | 3 |
| K1 | 3 | 1 | 3 | 3 |
| EQ39 (ToxoDB#54) | 3 | 1 | 3 | 3 |
| G13 | 3 | 1 | 3 | 3 |
| Strain | Time Post Infection | |
|---|---|---|
| 1 h | 3 h | |
| RH (ToxoDB#10) | + | + |
| BGD18 * (ToxoDB#1) | − | + |
| EQ40 | + | + |
| K1 | + | + |
| EQ39 (ToxoDB#54) | + | + |
| G13 | − | + |
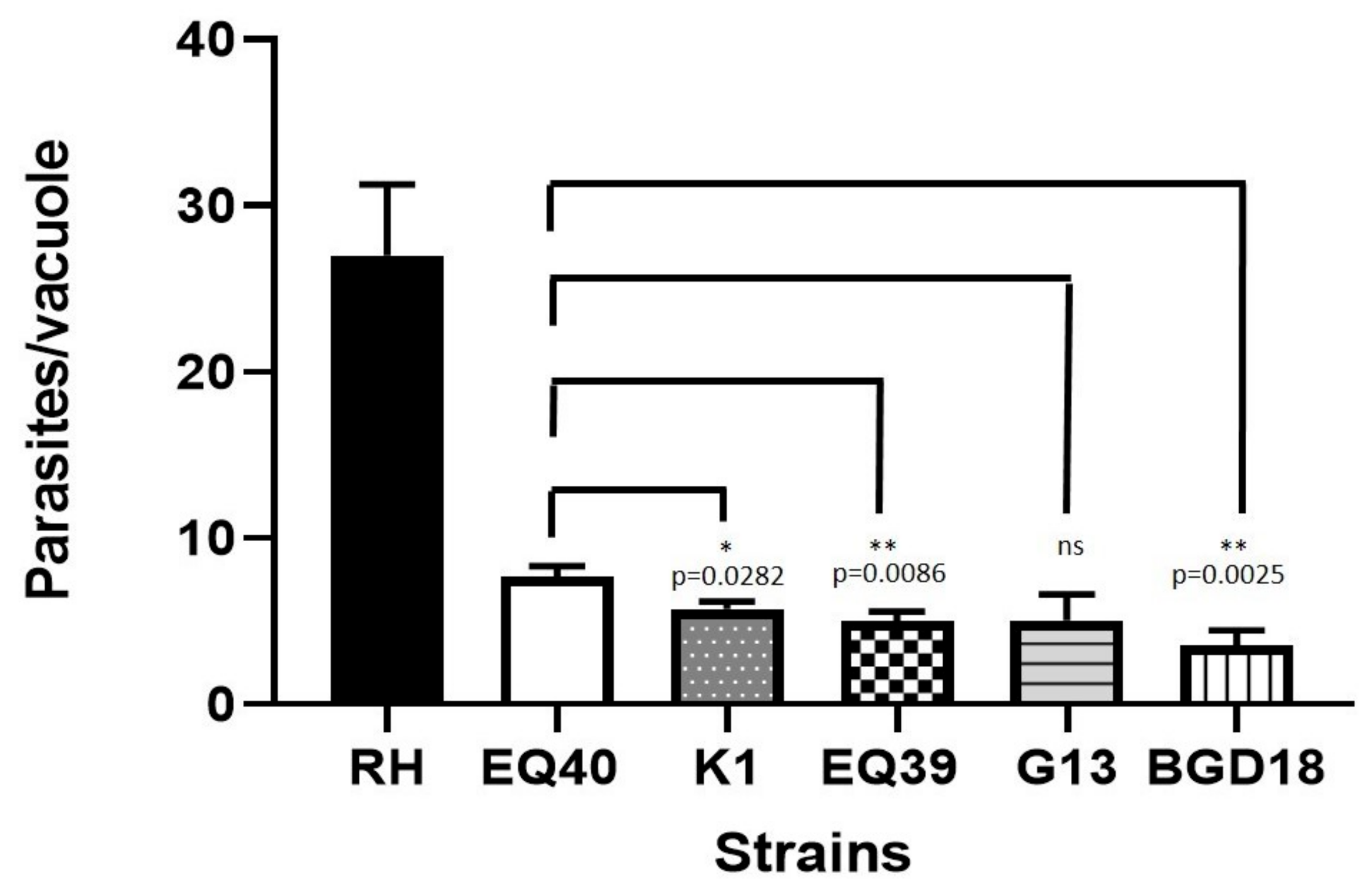
| Strain | Mean Growth Parameters 48 h Post Infection | |||
|---|---|---|---|---|
| No. of Parasites/Vacuole | SEM | No. of Divisions (log2 of Parasites/Vacuole) | SEM | |
| RH (ToxoDB#10) | 27 | 4.250 | 3.712 | 0.476 |
| BGD18 (ToxoDB#1) | 3.6 | 0.880 | 0.981 | 0.244 |
| EQ40 | 7.7 | 0.610 | 2.008 | 0.191 |
| K1 | 5.4 | 0.436 | 2.025 | 0.127 |
| EQ39 (ToxoDB#54) | 4.9 | 0.611 | 1.625 | 0.157 |
| G13 | 5.0 | 1.550 | 2.033 | 0.515 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uzelac, A.; Klun, I.; Ćirković, V.; Djurković-Djaković, O. In Vivo and In Vitro Virulence Analysis of Four Genetically Distinct Toxoplasma gondii Lineage III Isolates. Microorganisms 2020, 8, 1702. https://doi.org/10.3390/microorganisms8111702
Uzelac A, Klun I, Ćirković V, Djurković-Djaković O. In Vivo and In Vitro Virulence Analysis of Four Genetically Distinct Toxoplasma gondii Lineage III Isolates. Microorganisms. 2020; 8(11):1702. https://doi.org/10.3390/microorganisms8111702
Chicago/Turabian StyleUzelac, Aleksandra, Ivana Klun, Vladimir Ćirković, and Olgica Djurković-Djaković. 2020. "In Vivo and In Vitro Virulence Analysis of Four Genetically Distinct Toxoplasma gondii Lineage III Isolates" Microorganisms 8, no. 11: 1702. https://doi.org/10.3390/microorganisms8111702
APA StyleUzelac, A., Klun, I., Ćirković, V., & Djurković-Djaković, O. (2020). In Vivo and In Vitro Virulence Analysis of Four Genetically Distinct Toxoplasma gondii Lineage III Isolates. Microorganisms, 8(11), 1702. https://doi.org/10.3390/microorganisms8111702







