Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
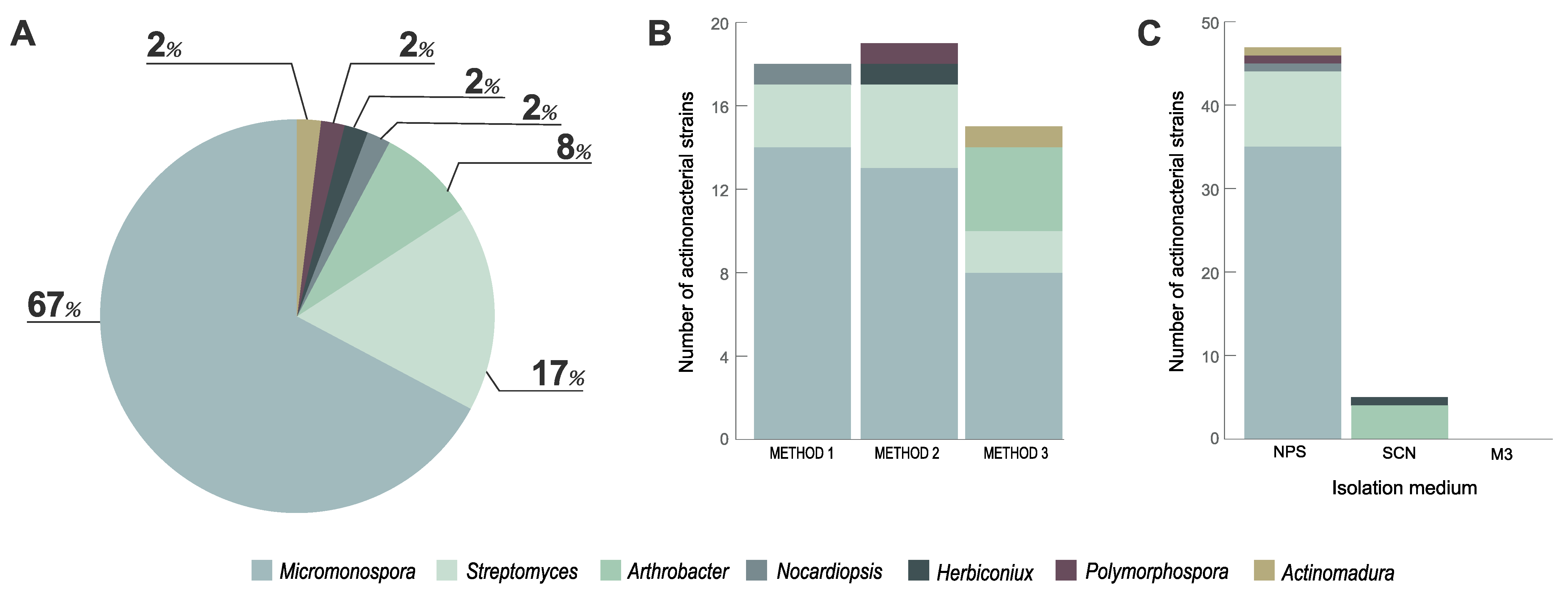
2.2. Isolation of Actinobacteria
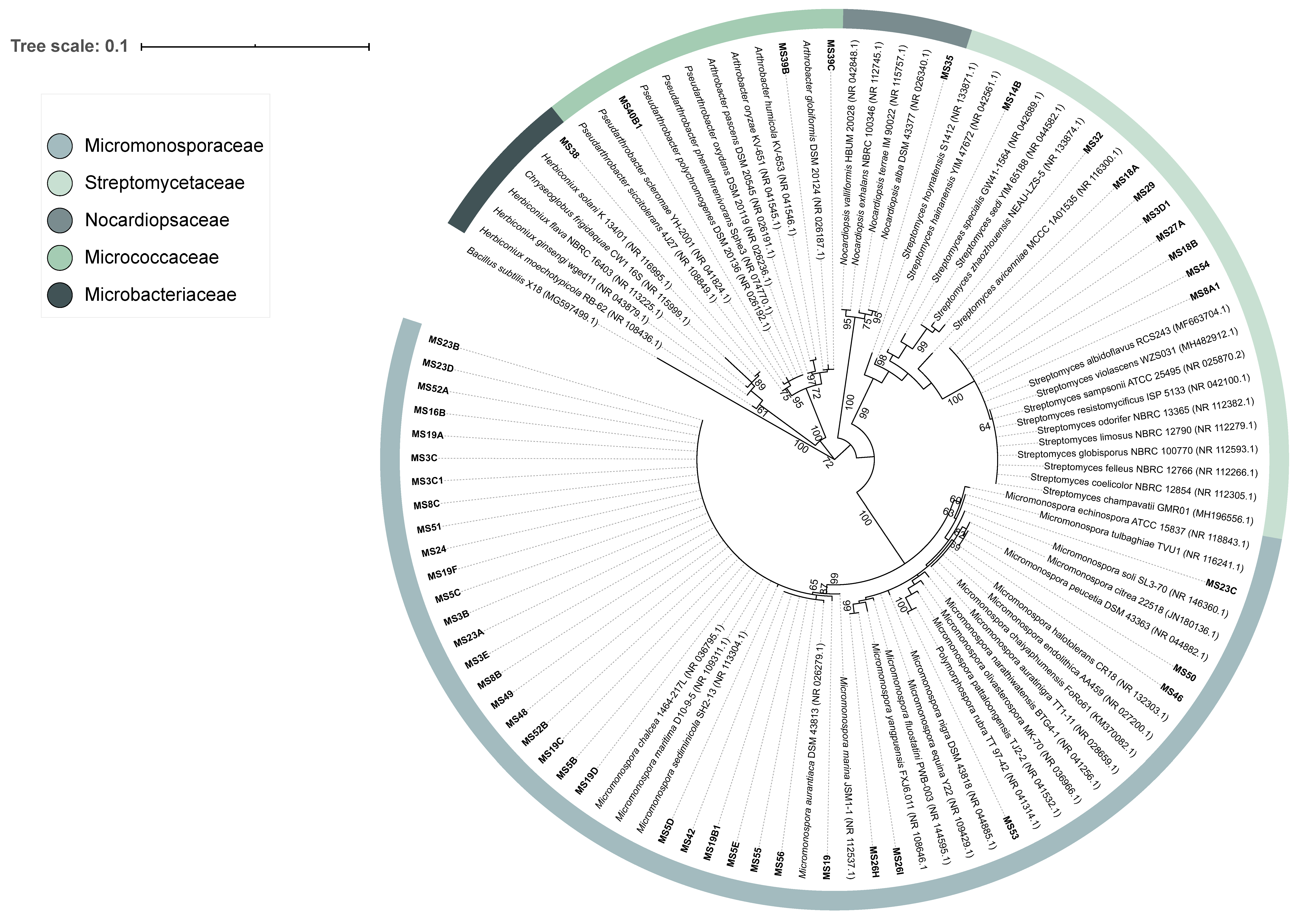
2.3. Identification of the Isolated Strains by 16S rRNA Gene Sequencing
2.4. Preparation of Crude Extracts
2.5. Antimicrobial Bioactivity Screening
2.6. Cytotoxic Bioactivity Screening
2.7. Statistical Analysis
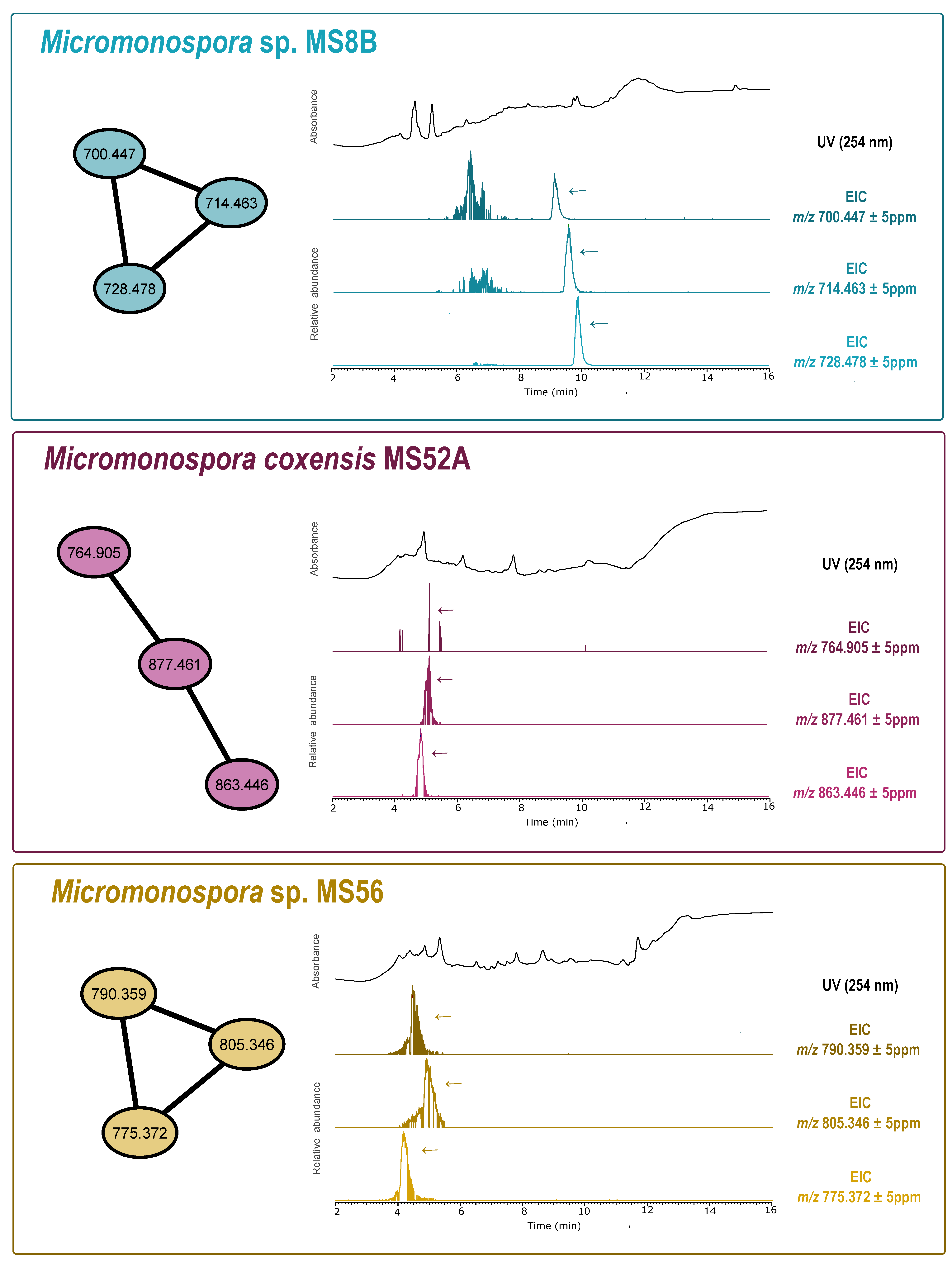
2.8. Dereplication and Molecular Network Analysis
3. Results
3.1. Actinobacteria Isolated from Marine Sediment
3.2. Bioactivity Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Duncan, K.R.; Haltli, B.; Gill, K.A.; Correa, H.; Berrue, F.; Kerr, R.G. Exploring the diversity and metabolic potential of actinomycetes from temperate marine sediments from Newfoundland, Canada. J. Ind. Microbiol. Biotechnol. 2015, 42, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Undabarrena, A.; Beltrametti, F.; Claverias, F.P.; Gonzalez, M.; Moore, E.R.; Seeger, M.; Camara, B. Exploring the Diversity and Antimicrobial Potential of Marine Actinobacteria from the Comau Fjord in Northern Patagonia, Chile. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Das, G. Advances in Pharmaceutical Biotechnology: Recent Progress and Future Applications, 1st ed.; Patra, J.K., Shukla, A.C., Das, G., Eds.; Springer: Berlin, Germany, 2020. [Google Scholar]
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. (Tokyo) 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Ward, A.C.; Bora, N. Diversity and biogeography of marine actinobacteria. Curr. Opin. Microbiol. 2006, 9, 279–286. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Marine actinobacterial metabolites: Current status and future perspectives. Microbiol. Res. 2013, 168, 311–332. [Google Scholar] [CrossRef]
- Prieto-Davo, A.; Dias, T.; Gomes, S.E.; Rodrigues, S.; Parera-Valadez, Y.; Borralho, P.M.; Pereira, F.; Rodrigues, C.M.; Santos-Sanches, I.; Gaudencio, S.P. The Madeira Archipelago As a Significant Source of Marine-Derived Actinomycete Diversity with Anticancer and Antimicrobial Potential. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Claverias, F.P.; Undabarrena, A.; Gonzalez, M.; Seeger, M.; Camara, B. Culturable diversity and antimicrobial activity of Actinobacteria from marine sediments in Valparaiso bay, Chile. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmao, L.; Amorim, A. Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 2010, 38, 203. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goodfellow, M. Nucleic Acid Techniques in Bacterial Systematics; Wiley: Hoboken, NJ, USA, 1991. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Mikheenko, A.; Garg, N.; Nothias, L.F.; Ninomiya, A.; Takada, K.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017, 13, 30–37. [Google Scholar] [CrossRef]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N. The natural products atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Weyland, H. Actinomycetes in North Sea and Atlantic Ocean sediments. Nature 1969, 223, 858. [Google Scholar] [CrossRef]
- Takizawa, M.; Colwell, R.R.; Hill, R.T. Isolation and diversity of actinomycetes in the chesapeake bay. Appl. Environ. Microbiol. 1993, 59, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Bredholt, H.; Fjærvik, E.; Johnsen, G.; Zotchev, S.B. Actinomycetes from Sediments in the Trondheim Fjord, Norway: Diversity and Biological Activity. Mar. Drugs 2008, 6, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.R.; Haltli, B.; Gill, K.A.; Kerr, R.G. Bioprospecting from Marine Sediments of New Brunswick, Canada: Exploring the Relationship between Total Bacterial Diversity and Actinobacteria Diversity. Mar. Drugs 2014, 12, 899–925. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.-Q.; Liu, Q.-X.; Pan, Z.-L.; Zhao, N.; Feng, Z.-X.; Wang, Y. Diversity and bioprospecting of culturable actinomycetes from marine sediment of the Yellow Sea, China. Arch. Microbiol. 2015, 197, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, M.S.; Elmallah, M.I.Y.; Hawas, U.W.; Abou El-Kassema, L.T.; Eid, M.A.G. Isolation and characterization of marine-derived actinomycetes with cytotoxic activity from the Red Sea coast. Asian Pac. J. Trop. Biomed. 2016, 6, 651–657. [Google Scholar] [CrossRef]
- Hames-Kocabas, E.E.; Uzel, A. Isolation strategies of marine-derived actinomycetes from sponge and sediment samples. J. Microbiol. Methods 2012, 88, 342–347. [Google Scholar] [CrossRef]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef]
- Jensen, P.R.; Gontang, E.; Mafnas, C.; Mincer, T.J.; Fenical, W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 2005, 7, 1039–1048. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Stach, J.E.; Pathom-aree, W.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie van Leeuwenhoek 2005, 87, 11–18. [Google Scholar] [CrossRef]
- Girão, M.; Ribeiro, I.; Ribeiro, T.; Azevedo, I.C.; Pereira, F.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Actinobacteria Isolated From Laminaria ochroleuca: A Source of New Bioactive Compounds. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Hassan, S.S.U.; Shaikh, A.L. Marine actinobacteria as a drug treasure house. Biomed. Pharmacother. 2017, 87, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Berdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; de la Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces zhaozhouensis. Mar. Drugs 2014, 13, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Daryamides A-C, weakly cytotoxic polyketides from a marine-derived actinomycete of the genus Streptomyces strain CNQ-085. J. Nat. Prod. 2006, 69, 1756–1759. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, D.; Thajuddin, N.; Panneerselvam, A. An antifungal compound: 4′ phenyl-1-napthyl–phenyl acetamide from Streptomyces sp. DPTB16. Facta Univ. Ser Med. Biol. 2008, 15, 7–12. [Google Scholar]
- McAlpine, J.B.; Banskota, A.H.; Charan, R.D.; Schlingmann, G.; Zazopoulos, E.; Piraee, M.; Janso, J.; Bernan, V.S.; Aouidate, M.; Farnet, C.M.; et al. Biosynthesis of diazepinomicin/ECO-4601, a Micromonospora secondary metabolite with a novel ring system. J. Nat. Prod. 2008, 71, 1585–1590. [Google Scholar] [CrossRef]
- Mullowney, M.; Ó hAinmhire, E.; Tanouye, U.; Burdette, J.; Pham, V.; Murphy, B. A Pimarane Diterpene and Cytotoxic Angucyclines from a Marine-Derived Micromonospora sp. in Vietnam’s East Sea. Mar. Drugs 2015, 13, 5815–5827. [Google Scholar] [CrossRef]
- Wang, H.; Yeo, S.L.; Xu, J.; Xu, X.; He, H.; Ronca, F.; Ting, A.E.; Wang, Y.; Yu, V.C.; Sim, M.M. Isolation of streptonigrin and its novel derivative from Micromonospora as inducing agents of p53-dependent cell apoptosis. J. Nat. Prod. 2002, 65, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.S.; Nicholson, B.; Teisan, S.; Lam, K.S.; Potts, B.C. Aureoverticillactam, a novel 22-atom macrocyclic lactam from the marine actinomycete Streptomyces aureoverticillatus. J. Nat. Prod. 2004, 67, 1400–1402. [Google Scholar] [CrossRef]
- Kwon, H.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “marinispora”. J. Am. Chem. Soc. 2006, 128, 1622–1632. [Google Scholar] [CrossRef]
- Lu, J.; Ma, Y.; Liang, J.; Xing, Y.; Xi, T.; Lu, Y. Aureolic acids from a marine-derived Streptomyces sp. WBF16. Microbiol. Res. 2012, 167, 590–595. [Google Scholar] [CrossRef]
- Han, X.-X.; Cui, C.-B.; Gu, Q.-Q.; Zhu, W.-M.; Liu, H.-B.; Gu, J.-Y.; Osada, H. ZHD-0501, a novel naturally occurring staurosporine analog from Actinomadura sp. 007. Tetrahedron. Lett. 2005, 46, 6137–6140. [Google Scholar] [CrossRef]
- Maskey, R.P.; Li, F.; Qin, S.; Fiebig, H.H.; Laatsch, H. Chandrananimycins A approximately C: Production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J. Antibiot. (Tokyo) 2003, 56, 622–629. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, X.; Kim, S.J.; Zhang, W. Antimycin-type depsipeptides: Discovery, biosynthesis, chemical synthesis, and bioactivities. Nat. Prod. Rep. 2016, 33, 1146–1165. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, C.; Liu, C.; Ju, J.; Ma, J. Genome Sequencing of Streptomyces atratus SCSIOZH16 and Activation Production of Nocardamine via Metabolic Engineering. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Song, Y.-N.; Zhang, W.-J.; Bi, S.-F.; Jiao, R.-H.; Tan, R.-X.; Ge, H.-M. New ansamycin analogues from the mutant strain of Streptomyces seoulensis. J Antibiot. B 2015, 68, 757–759. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore production by actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Das, P.; Sivapathasekaran, C.; Sen, R. Antimicrobial biosurfactants from marine Bacillus circulans: Extracellular synthesis and purification. Lett. Appl. Microbiol. 2009, 48, 281–288. [Google Scholar] [CrossRef]
- Desriac, F.; Jégou, C.; Balnois, E.; Brillet, B.; Chevalier, P.L.; Fleury, Y. Antimicrobial Peptides from Marine Proteobacteria. Mar. Drugs 2013, 11, 3632–3660. [Google Scholar] [CrossRef] [PubMed]
- Kjaerulff, L.; Nielsen, A.; Mansson, M.; Gram, L.; Larsen, T.O.; Ingmer, H.; Gotfredsen, C.H. Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar. Drugs 2013, 11, 5051–5062. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-W.; Xu, L.; Mierzwa, R.; He, L.; Terracciano, J.; Patel, M.; Gullo, V.; Black, T.; Zhao, W.; Chan, T.-M.; et al. Two novel antibiotics, Sch 419558 and Sch 419559, produced by Pseudomonas fluorescens: Effect on activity by overexpression of RpoE. Bioorg. Med. Chem. 2004, 12, 3333–3338. [Google Scholar] [CrossRef]
- Dijkstra, J.; van der Laan, T.; Akkerman, O.; Bolhuis, M.; De Lange, W.; Kosterink, J.G.; van der Werf, T.S.; Alffenaar, J.-W.C.; Van Soolingen, D. In vitro susceptibility of Mycobacterium tuberculosis to amikacin, kanamycin, and capreomycin. Antimicrob. Agents Chemother. 2018, 62, e01724-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Kim, J.D.; Hong, J.S.; Ham, J.H.; Kim, B.S. Identification of antifungal niphimycin from Streptomyces sp. KP6107 by screening based on adenylate kinase assay. J. Basic. Microbiol. 2013, 53, 581–589. [Google Scholar] [CrossRef]

| Isolate | Closest Identification | Disk Diffusion Method | |||||
|---|---|---|---|---|---|---|---|
| E. coli | S. typhimurium | S. aureus | B. subtilis | C. albicans | MIC (µg mL−1) | ||
| MS8A1 | Streptomyces sp. |  |  |  |  |  | 15.62 |
| MS18A | Streptomyces sp. |  |  |  |  |  | 15.62 |
| MS18B | Streptomyces sp. |  |  |  |  |  | 3.90 |
| MS27A | Streptomyces sp. |  |  |  |  |  | 62.5 |
| MS29 | Streptomyces sp. |  |  |  |  |  | 15.62 |
| MS54 | Streptomyces sp. |  |  |  |  |  | 125 |
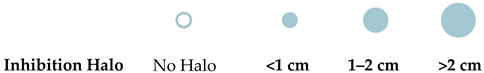
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, I.; Girão, M.; Alexandrino, D.A.M.; Ribeiro, T.; Santos, C.; Pereira, F.; Mucha, A.P.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal. Microorganisms 2020, 8, 1691. https://doi.org/10.3390/microorganisms8111691
Ribeiro I, Girão M, Alexandrino DAM, Ribeiro T, Santos C, Pereira F, Mucha AP, Urbatzka R, Leão PN, Carvalho MF. Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal. Microorganisms. 2020; 8(11):1691. https://doi.org/10.3390/microorganisms8111691
Chicago/Turabian StyleRibeiro, Inês, Mariana Girão, Diogo A. M. Alexandrino, Tiago Ribeiro, Chiara Santos, Filipe Pereira, Ana P. Mucha, Ralph Urbatzka, Pedro N. Leão, and Maria F. Carvalho. 2020. "Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal" Microorganisms 8, no. 11: 1691. https://doi.org/10.3390/microorganisms8111691
APA StyleRibeiro, I., Girão, M., Alexandrino, D. A. M., Ribeiro, T., Santos, C., Pereira, F., Mucha, A. P., Urbatzka, R., Leão, P. N., & Carvalho, M. F. (2020). Diversity and Bioactive Potential of Actinobacteria Isolated from a Coastal Marine Sediment in Northern Portugal. Microorganisms, 8(11), 1691. https://doi.org/10.3390/microorganisms8111691








