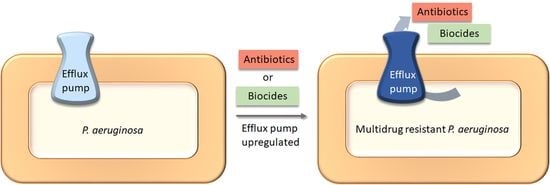
Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source and Identification of P. aeruginosa Isolates
2.2. Antimicrobial Susceptibility Testing
Establishing Biocide Epidemiologic Cut-off Values
2.3. Efflux Pump Inhibition Using a Checkerboard Assay
2.4. Gene Expression Analysis by Reverse-Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
2.5. DNA Extraction and Whole Genome Sequencing (WGS)
2.6. WGS Assembly, Annotation and Analysis
2.7. Genome Accession Numbers
2.8. Statistical Analysis
3. Results
3.1. P. aeruginosa Isolates from Wastewater Display a High Prevalence for Multidrug Resistance in Contrast to Clinical and Veterinary Isolates
3.2. P. aeruginosa Has a High Resistance against Common Biocides Used in Clinical Settings
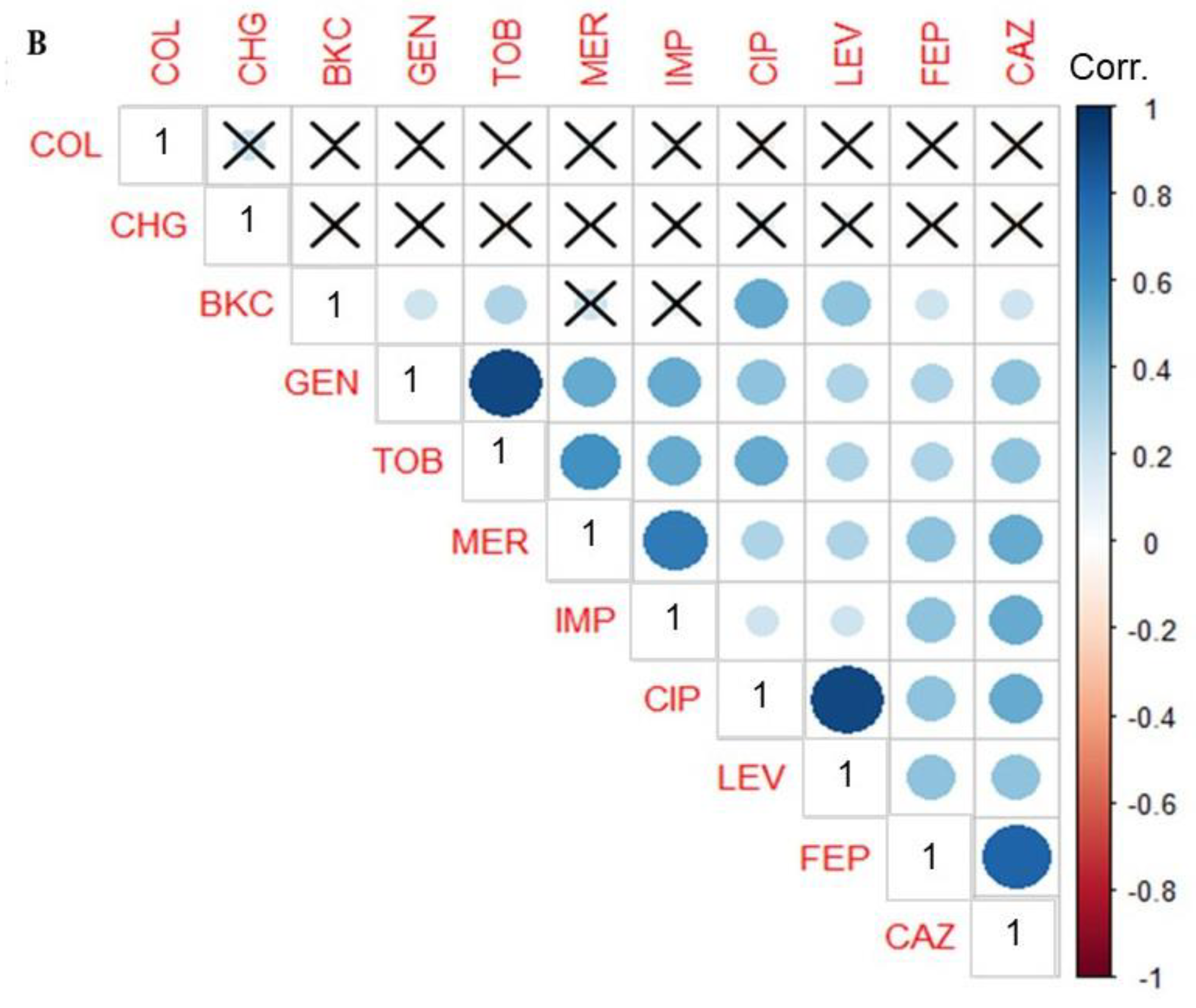
3.3. Phenotypic Correlation between Biocide and Antimicrobial Resistance in P. aeruginosa Isolates
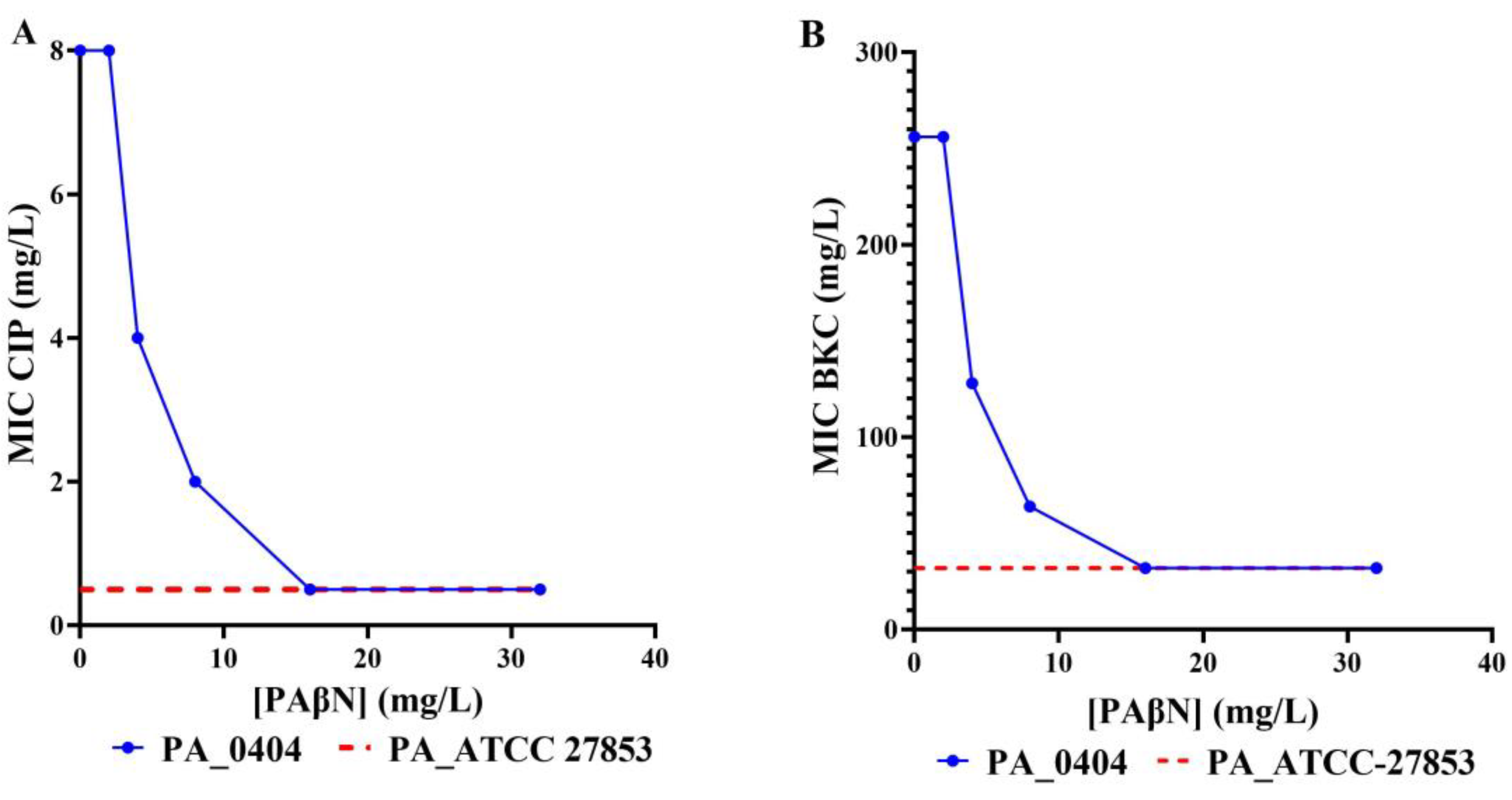
3.4. Efflux Pump Inhibition Reveals RND Pump Mediated Biocide and Antimicrobial Resistance
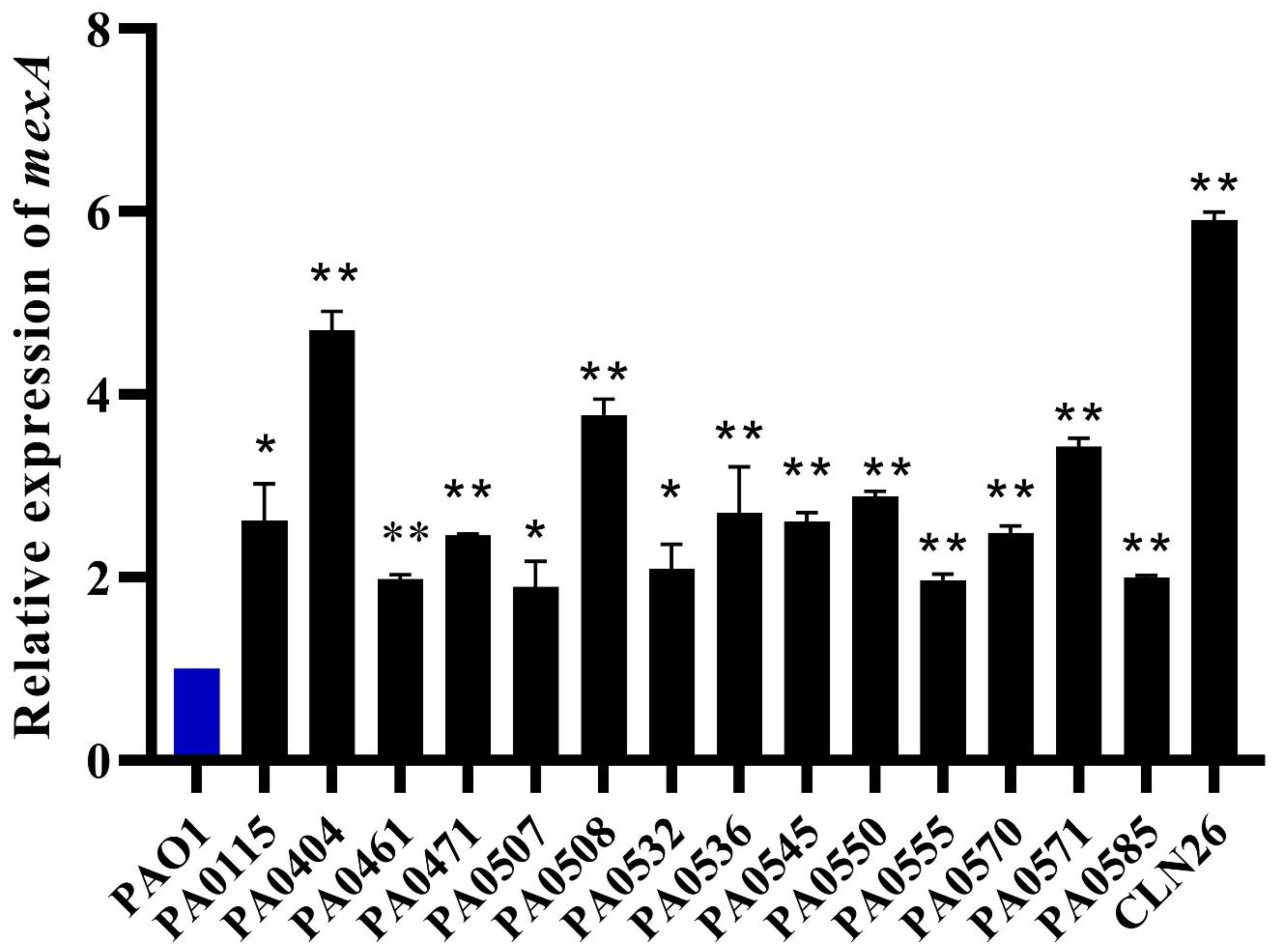
3.5. RT-qPCR Revealed MexAB-OprM efflux Pump Overexpression
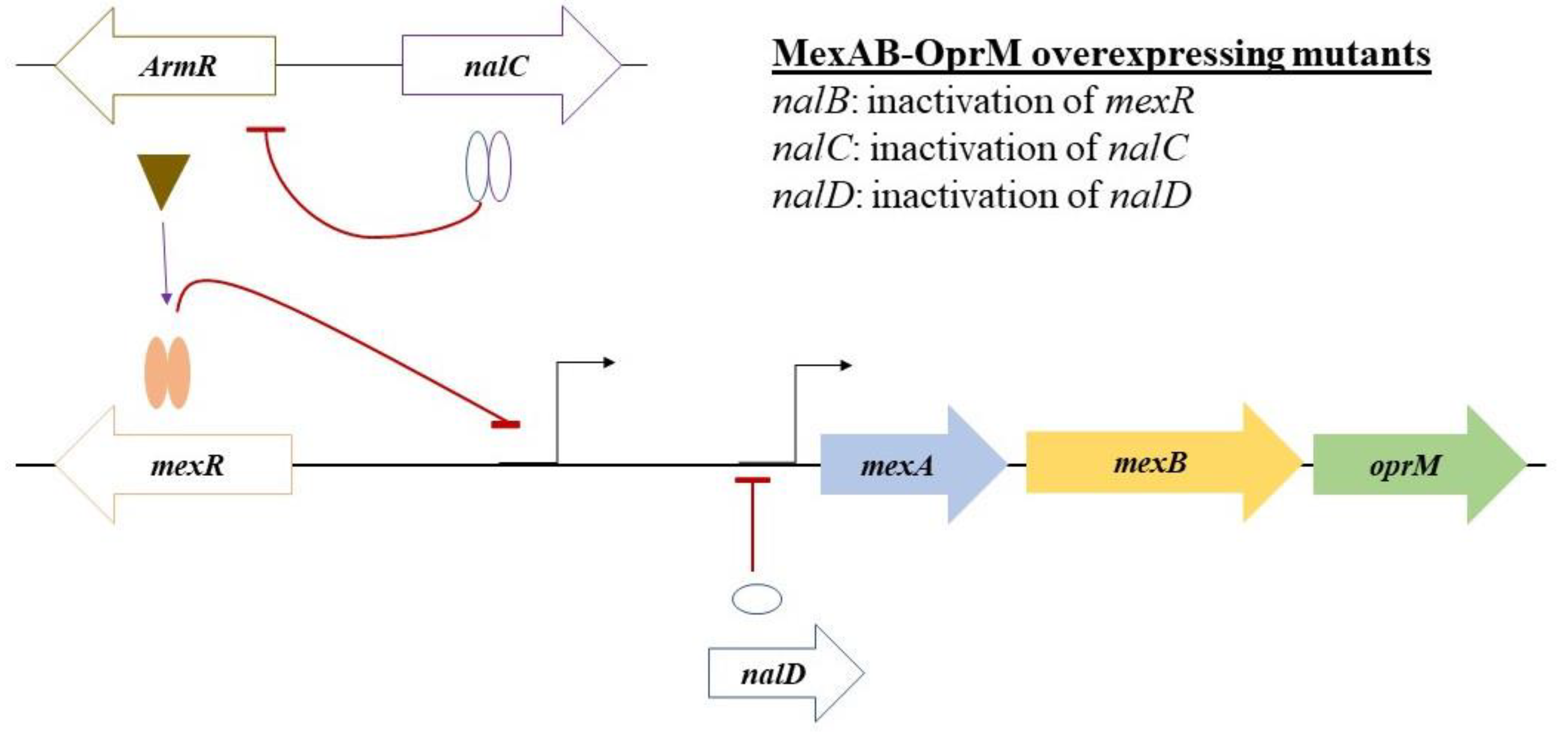
3.6. WGS Analysis Reveals Multiple Mutations in the Regulators of the mexab-oprM Efflux Pump
3.7. The Role of Other Resistance Gene Determinants in the Development of High-Level Resistance to FQs and Biocides
3.8. The Pangenome Analysis Indicated an Evolutionary Divergence between mexAB-oprM Efflux Pump Overexpressing P. aeruginosa Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Snyder, L.; Loman, N.; Faraj, L.; Levi, K.; Weinstock, J.; Boswell, T.; Pallen, M.; Ala’Aldeen, D. Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Eurosurveillance 2013, 18, 42. [Google Scholar] [CrossRef] [Green Version]
- Kumarage, J.; Khonyongwa, K.; Khan, A.; Desai, N.; Hoffman, P.; Taori, S. Transmission of multi-drug resistant Pseudomonas aeruginosa between two flexible ureteroscopes and an outbreak of urinary tract infection: The fragility of endoscope decontamination. J. Hosp. Infect. 2019, 102, 89–94. [Google Scholar] [CrossRef]
- Slekovec, C.; Plantin, J.; Cholley, P.; Thouverez, M.; Talon, D.; Bertrand, X.; Hocquet, D. Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS ONE 2012, 7, e49300. [Google Scholar] [CrossRef] [Green Version]
- Quick, J.; Cumley, N.; Wearn, C.M.; Niebel, M.; Constantinidou, C.; Thomas, C.M.; Pallen, M.J.; Moiemen, N.S.; Bamford, A.; Oppenheim, B. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open 2014, 4, e006278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tümmler, B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampf, G. Acquired resistance to chlorhexidine–Is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 2016, 94, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa–Mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 100640. [Google Scholar] [CrossRef] [PubMed]
- Treepong, P.; Kos, V.; Guyeux, C.; Blanc, D.; Bertrand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cheng, A.C.; Turnidge, J.; Collignon, P.; Looke, D.; Barton, M.; Gottlieb, T. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg. Infect. Dis. 2012, 18, 1453. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Granrot, A.; Ahl, J.; Tham, J.; Resman, F.; Riesbeck, K.; Månsson, F. Antimicrobial combination treatment including ciprofloxacin decreased the mortality rate of Pseudomonas aeruginosa bacteraemia: A retrospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1187–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kümmerer, K.; Al-Ahmad, A.; Mersch-Sundermann, V. Biodegradability of some antibiotics, elimination of the genotoxicity and affection of wastewater bacteria in a simple test. Chemosphere 2000, 40, 701–710. [Google Scholar] [CrossRef]
- Liu, W.-L.; Chang, P.-C.; Chen, Y.-Y.; Lai, C.-C. Impact of fluoroquinolone consumption on resistance of healthcare-associated Pseudomonas aeruginosa. J. Infect. 2012, 64, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Chen, Y.; Jiang, S.; Shen, P.; Lu, X.; Xiao, Y. Association between the rate of fluoroquinolones-resistant gram-negative bacteria and antibiotic consumption from China based on 145 tertiary hospitals data in 2014. BMC Infect. Dis. 2020, 20, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tandukar, M.; Oh, S.; Tezel, U.; Konstantinidis, K.T.; Pavlostathis, S.G. Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ. Sci. Technol. 2013, 47, 9730–9738. [Google Scholar] [CrossRef]
- Kim, M.; Weigand, M.R.; Oh, S.; Hatt, J.K.; Krishnan, R.; Tezel, U.; Pavlostathis, S.G.; Konstantinidis, K.T. Widely used benzalkonium chloride disinfectants can promote antibiotic resistance. Appl. Environ. Microbiol. 2018, 84, e01201-18. [Google Scholar] [CrossRef] [Green Version]
- Health Council of the Netherlands. Resistance due to Disinfectants, Background Report to the Advisory Report Careful Use of Disinfectants. The Hague: Health Council of the Netherlands; Publication No. A16/03E. 2016. Available online: www.healthcouncil.nl. (accessed on 22 October 2020).
- Pereira, B.M.P.; Tagkopoulos, I. Benzalkonium chlorides: Uses, regulatory status, and microbial resistance. Appl. Environ. Microbiol. 2019, 85, e00377-19. [Google Scholar] [CrossRef] [Green Version]
- Marple, B.; Roland, P.; Benninger, M. Safety review of benzalkonium chloride used as a preservative in intranasal solutions: An overview of conflicting data and opinions. Otolaryngol. -Head Neck Surg. 2004, 130, 131–141. [Google Scholar] [CrossRef]
- Weatherly, L.M.; Gosse, J.A. Triclosan exposure, transformation, and human health effects. J. Toxicol. Environ. Health B 2017, 20, 447–469. [Google Scholar] [CrossRef]
- Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Comparing the selective and co-selective effects of different antimicrobials in bacterial communities. Int. J. Antimicrob. Agents 2019, 53, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.R.; Kappell, A.D.; McNamara, P.J. Benzalkonium chloride alters phenotypic and genotypic antibiotic resistance profiles in a source water used for drinking water treatment. Environ. Pollut. 2020, 257, 113472. [Google Scholar] [CrossRef]
- Mc Cay, P.H.; Ocampo-Sosa, A.A.; Fleming, G.T. Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 2010, 156, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, D.E.; McNamara, P.J. The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 2015, 5, 780. [Google Scholar] [CrossRef] [Green Version]
- Venter, H.; Henningsen, M.L.; Begg, S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017, 61, 1–10. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Pseudomonas aeruginosa: Resistance to the max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef] [Green Version]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar]
- Dreier, J.; Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [Green Version]
- Sobel, M.L.; Hocquet, D.; Cao, L.; Plesiat, P.; Poole, K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 1782–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12. [Google Scholar] [CrossRef] [Green Version]
- Rehman, A.; Patrick, W.M.; Lamont, I.L. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 2019, 68, 1–10. [Google Scholar] [CrossRef]
- Bruchmann, S.; Dötsch, A.; Nouri, B.; Chaberny, I.F.; Häussler, S. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 2013, 57, 1361–1368. [Google Scholar] [CrossRef] [Green Version]
- Rozman, U.; Duh, D.; Cimerman, M.; Turk, S.Š. Hospital wastewater effluent: Hot spot for antibiotic resistant bacteria. J. Watersanitation Hyg. Dev. 2020, 10, 171–178. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef]
- Lien, L.T.Q.; Hoa, N.Q.; Chuc, N.T.K.; Thoa, N.T.M.; Phuc, H.D.; Diwan, V.; Dat, N.T.; Tamhankar, A.J.; Lundborg, C.S. Antibiotics in wastewater of a rural and an urban hospital before and after wastewater treatment, and the relationship with antibiotic use—a one year study from Vietnam. Int. J. Environ. Res. Public Health 2016, 13, 588. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Lin, J.; Ma, J.; Cronan, J.E.; Wang, H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 2010, 54, 689–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampf, G. Adaptive microbial response to low-level benzalkonium chloride exposure. J. Hosp. Infect. 2018, 100, e1–e22. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front. Cell. Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Mowla, R.; Ji, S.; Guo, L.; Lopes, M.A.D.B.; Jin, C.; Song, D.; Ma, S.; Venter, H. Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 2018, 143, 699–709. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Alenzy, R.; Song, D.; Liu, X.; Teng, Y.; Mowla, R.; Ma, Y.; Polyak, S.W.; Venter, H.; Ma, S. Structural optimization of natural product nordihydroguaretic acid to discover novel analogues as AcrB inhibitors. Eur. J. Med. Chem. 2020, 186, 111910. [Google Scholar] [CrossRef]
- Pournaras, S.; Maniati, M.; Spanakis, N.; Ikonomidis, A.; Tassios, P.; Tsakris, A.; Legakis, N.; Maniatis, A. Spread of efflux pump-overexpressing, non-metallo-β-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with bla VIM endemicity. J. Antimicrob. Chemother. 2005, 56, 761–764. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [Green Version]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, 566–573. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Volume 1, p. 409. [Google Scholar]
- Hirakata, Y.; Srikumar, R.; Poole, K.; Gotoh, N.; Suematsu, T.; Kohno, S.; Kamihira, S.; Hancock, R.E.; Speert, D.P. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 2002, 196, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Coombs, G.B.J.; Daley, D.; Collignon, P.; Cooley, L.; Gottlieb, T.; Iredell, J.; Kotsanas, D.; Nimmo, G.; Robson, J.; On behalf of the Australian Group on Antimicrobial Resistance and Australian Commission on Safety and Quality in Health Care. Australian Group on Antimicrobial Resistance Sepsis Outcomes Programs: 2018 Report. Sydney: ACSQHC. Available online: https://www.safetyandquality.gov.au/publications-and-resources/resource-library/agar-sepsis-outcome-programs-2018-report (accessed on 22 October 2020).
- ECDC. Surveillance of Antimicrobial Resistance in Europe 2018. Stockholm: ECDC. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2018 (accessed on 22 October 2020).
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Bondurant, S.W.; Duley, C.M.; Harbell, J.W. Demonstrating the persistent antibacterial efficacy of a hand sanitizer containing benzalkonium chloride on human skin at 1, 2, and 4 h after application. Am. J. Infect. Control 2019, 47, 928–932. [Google Scholar] [CrossRef] [Green Version]
- Kraupner, N.; Ebmeyer, S.; Bengtsson-Palme, J.; Fick, J.; Kristiansson, E.; Flach, C.-F.; Larsson, D.J. Selective concentration for ciprofloxacin resistance in Escherichia coli grown in complex aquatic bacterial biofilms. Environ. Int. 2018, 116, 255–268. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. [Google Scholar] [CrossRef] [Green Version]
- Tetard, A.; Zedet, A.; Girard, C.; Plésiat, P.; Llanes, C. Cinnamaldehyde induces expression of efflux pumps and multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01081-19. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Hatt, J.K.; Weigand, M.R.; Krishnan, R.; Pavlostathis, S.G.; Konstantinidis, K.T. Genomic and transcriptomic insights into how bacteria withstand high concentrations of benzalkonium chloride biocides. Appl. Environ. Microbiol. 2018, 84, e00197-18. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Jin, M.; Nguyen, S.H.; Mao, L.; Li, J.; Coin, L.J.; Yuan, Z.; Guo, J. Non-antibiotic antimicrobial triclosan induces multiple antibiotic resistance through genetic mutation. Environ. Int. 2018, 118, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Grosso, F.; Quinteira, S.; Brilhante, M.; Ramos, H.; Peixe, L. Two decades of bla VIM-2-producing Pseudomonas aeruginosa dissemination: An interplay between mobile genetic elements and successful clones. J. Antimicrob. Chemother. 2018, 73, 873–882. [Google Scholar] [CrossRef] [Green Version]
- Chávez-Jacobo, V.M.; Hernández-Ramírez, K.C.; Romo-Rodríguez, P.; Pérez-Gallardo, R.V.; Campos-García, J.; Gutiérrez-Corona, J.F.; García-Merinos, J.P.; Meza-Carmen, V.; Silva-Sánchez, J.; Ramírez-Díaz, M.I. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents Chemother. 2018, 62, e02629-17. [Google Scholar]
- López-Causapé, C.; Sommer, L.M.; Cabot, G.; Rubio, R.; Ocampo-Sosa, A.A.; Johansen, H.K.; Figuerola, J.; Cantón, R.; Kidd, T.J.; Molin, S. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci. Rep. 2017, 7, 5555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocampo-Sosa, A.A.; Fernández-Martínez, M.; Cabot, G.; Peña, C.; Tubau, F.; Oliver, A.; Martínez-Martínez, L. Draft genome sequence of the quorum-sensing and biofilm-producing Pseudomonas aeruginosa strain Pae221, belonging to the epidemic high-risk clone sequence type 274. Genome Announc. 2015, 3, e01343-14. [Google Scholar] [CrossRef] [Green Version]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, L.J. Bacterial biocide resistance: A new scourge of the infectious disease world? Arch. Dis. Child. 2019, 104, 1029–1033. [Google Scholar] [CrossRef] [PubMed]


| Strains | ST | Antimicrobial Susceptibility | RP | MexAB-OprM | Fluoroquinolone Resistance Determinants | Mutation in MexAB-OprM Efflux Pump Regulators | Triclosan | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEP | CAZ | CIP | LEV | GEN | TOB | MER | IMP | COL | GyrA | GyrB | ParC | ParE | CrpP | MexR | NalC | NalD | FabV | ||||
| PAO1 | S | S | S | S | S | S | S | S | S | S | 1 | ||||||||||
| PA0115 | 235 | R | R | R | R | R | S | S | S | S | MDR | 2.622 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0404 | 235 | R | R | R | R | R | R | S | R | S | MDR | 4.705 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | T11N | P36L |
| PA0461 | 235 | R | R | R | R | R | R | R | R | S | MDR | 1.986 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0471 | 235 | R | R | R | R | S | S | R | R | S | MDR | 2.462 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0507 | 235 | R | R | R | R | R | S | S | S | S | MDR | 1.896 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0508 | 235 | R | R | R | R | R | R | S | S | S | MDR | 3.778 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0532 | 235 | R | R | R | R | R | R | R | R | S | MDR | 2.097 | T83I | − | S87L | D533E | − | V126E | S209R, G71E | − | P36L |
| PA0536 | 815 | R | R | R | R | R | R | R | R | S | MDR | 2.707 | D87N | − | V297I | D533E | + | V126E | S209R, G71E | − | P36L |
| PA0545 | 815 | R | R | R | R | R | R | R | R | S | MDR | 2.610 | D87N | − | V297I | D533E | + | V126E | S209R, G71E | − | P36L |
| PA0550 | 235 | R | R | R | R | R | S | S | S | S | MDR | 2.882 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0555 | 235 | R | R | R | R | S | S | S | S | S | NMDR | 1.969 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0570 | 235 | R | R | R | R | R | S | S | S | S | MDR | 2.486 | T83I | − | S87L, TR* | D533E | − | V126E | S209R, G71E, E153Q, TR*** | − | P36L |
| PA0571 | 235 | R | R | R | R | R | S | S | S | S | MDR | 3.427 | T83I | − | S87L | D533EE569-** | − | V126E | S209R, G71E, E153Q | − | P36L |
| PA0585 | 235 | R | R | R | R | R | S | S | S | S | MDR | 1.996 | T83I | − | S87L | D533E | − | V126E | S209R, G71E, E153Q | − | P36L |
| CLN_26 | 274 | R | R | R | R | S | S | S | S | S | NMDR | 5.911 | T83I | E468D, H148N | − | P438S, L501F | − | − | S209R, G71E | − | P260T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amsalu, A.; Sapula, S.A.; De Barros Lopes, M.; Hart, B.J.; Nguyen, A.H.; Drigo, B.; Turnidge, J.; Leong, L.E.; Venter, H. Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms 2020, 8, 1647. https://doi.org/10.3390/microorganisms8111647
Amsalu A, Sapula SA, De Barros Lopes M, Hart BJ, Nguyen AH, Drigo B, Turnidge J, Leong LE, Venter H. Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms. 2020; 8(11):1647. https://doi.org/10.3390/microorganisms8111647
Chicago/Turabian StyleAmsalu, Anteneh, Sylvia A. Sapula, Miguel De Barros Lopes, Bradley J. Hart, Anh H. Nguyen, Barbara Drigo, John Turnidge, Lex EX Leong, and Henrietta Venter. 2020. "Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots" Microorganisms 8, no. 11: 1647. https://doi.org/10.3390/microorganisms8111647
APA StyleAmsalu, A., Sapula, S. A., De Barros Lopes, M., Hart, B. J., Nguyen, A. H., Drigo, B., Turnidge, J., Leong, L. E., & Venter, H. (2020). Efflux Pump-Driven Antibiotic and Biocide Cross-Resistance in Pseudomonas aeruginosa Isolated from Different Ecological Niches: A Case Study in the Development of Multidrug Resistance in Environmental Hotspots. Microorganisms, 8(11), 1647. https://doi.org/10.3390/microorganisms8111647