Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water
Abstract
1. Introduction
2. Materials and Methods
2.1. Resistance Testing and Preparation of Inoculation Cultures
2.2. Lettuce-in-Pot: Growth Conditions and Inoculation
2.2.1. Setup
2.2.2. Water-Inoculation Experiments
2.2.3. Manure-Inoculation Experiment
2.3. Sampling and Culture-Based Approach
2.3.1. Starting Materials and Sampling Scheme
2.3.2. Lettuce Leave Sampling
2.3.3. Soil Sampling
2.4. ARG Detection in Re-Isolated E. coli pEK499 and Secondary MDR Colonies
2.5. Molecular Detection of blaCTX-M-15 in Plant and Soil
2.6. Statistics
3. Results
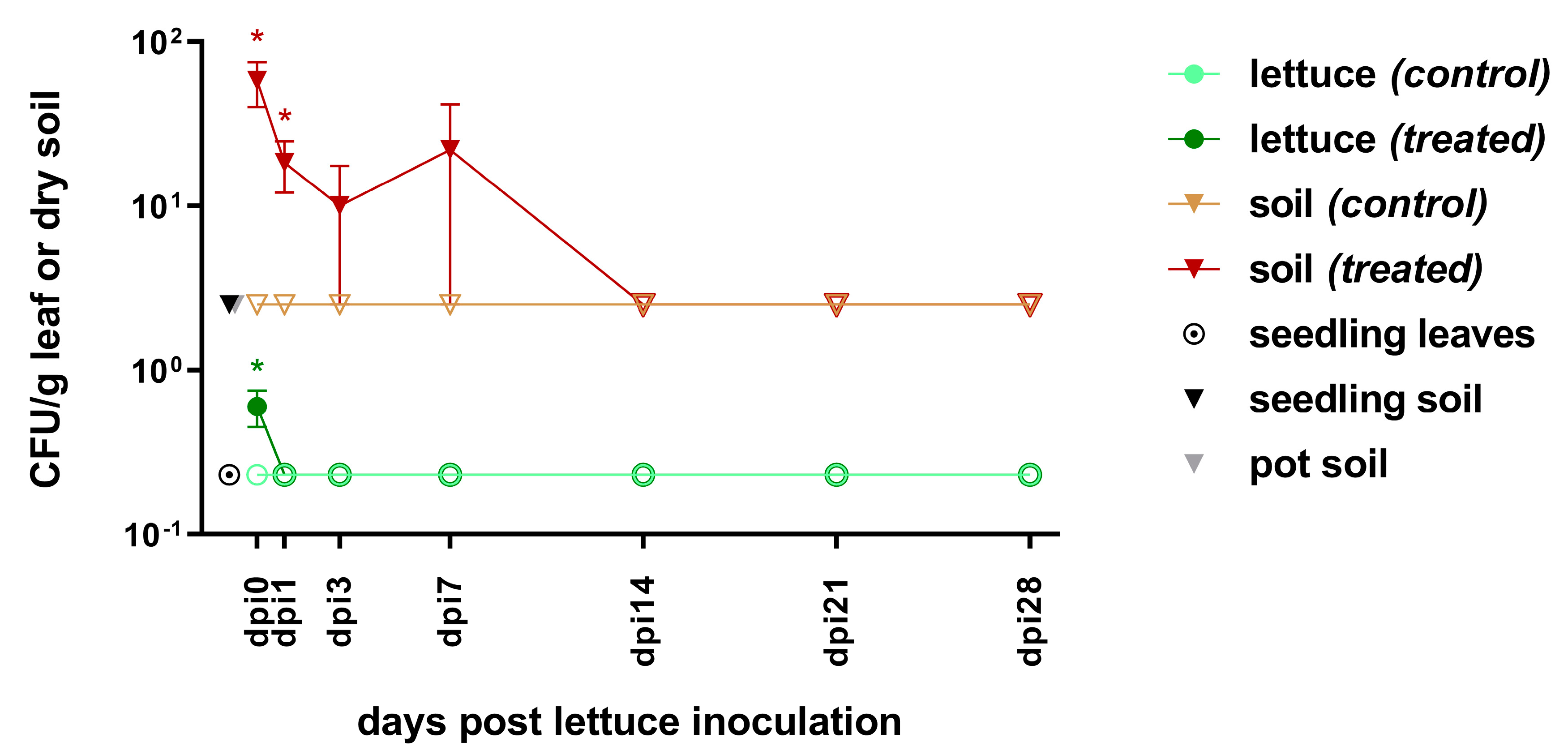
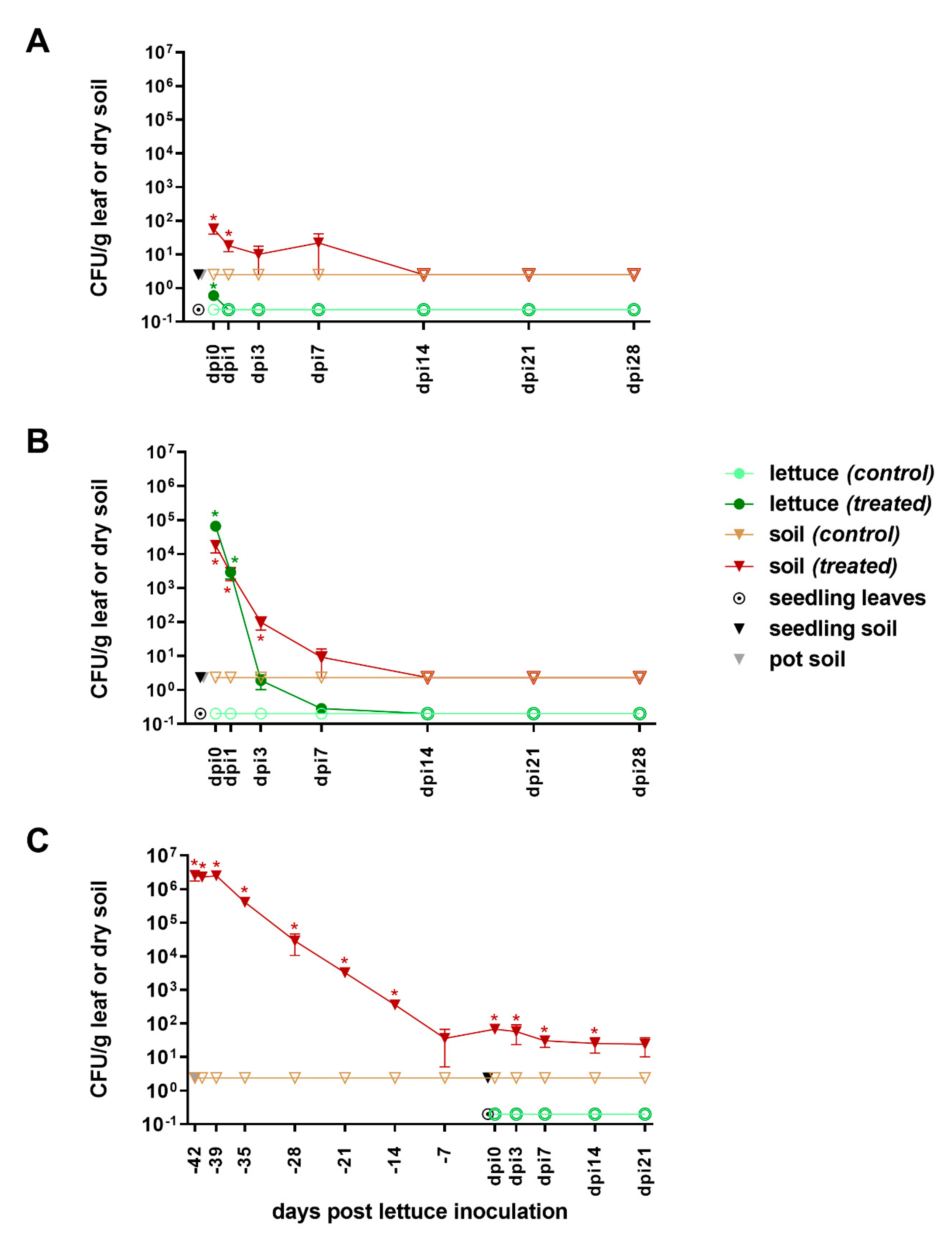
3.1. Survival of E. coli pEK499 in Lettuce-in-Pot System
3.1.1. Pre-Inoculation
3.1.2. From Inoculation to Harvest
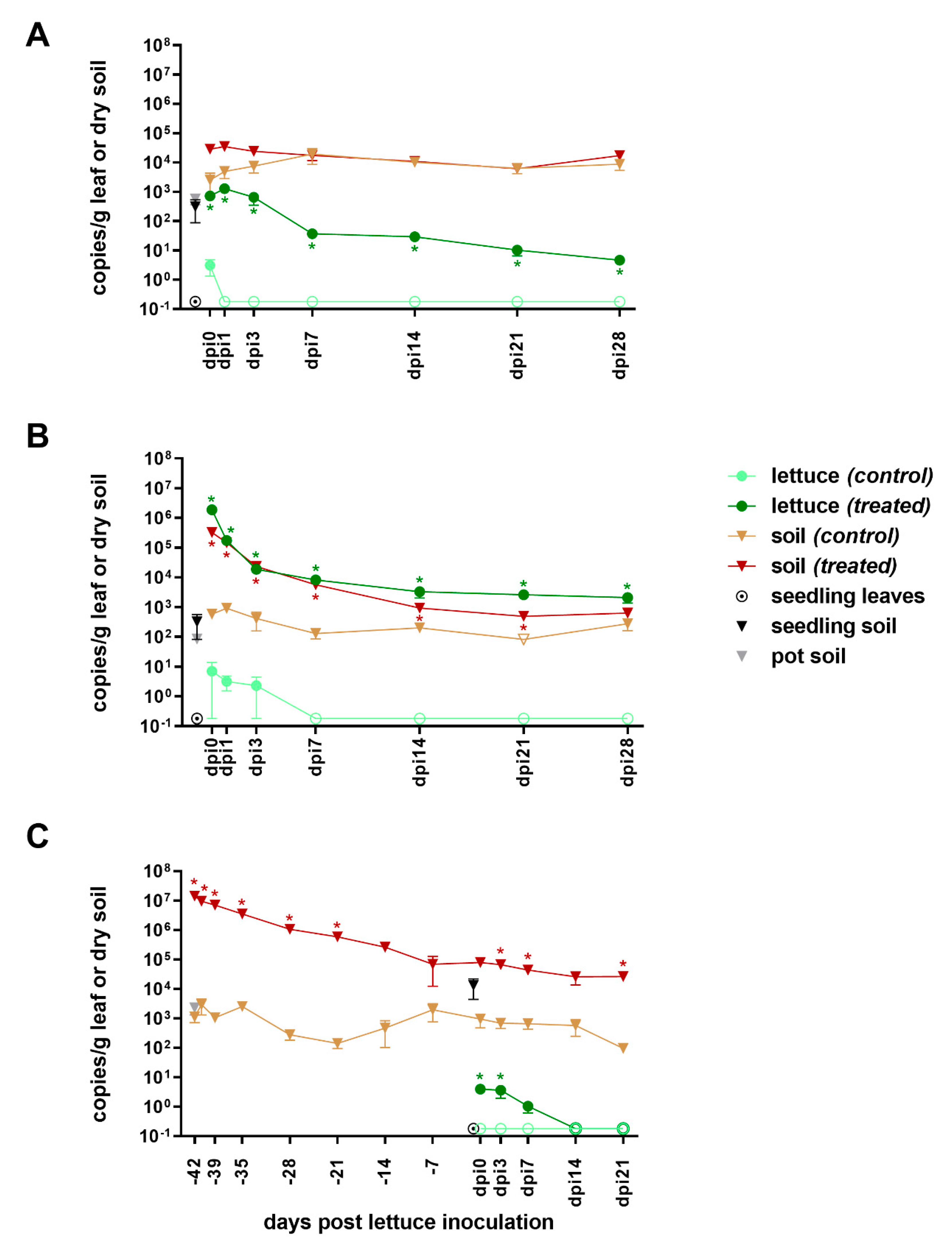
3.2. Detection of blaCTX-M-15 in the Lettuce-in-Pot System
3.2.1. Pre-Inoculation
3.2.2. From Inoculation to Harvest
3.3. ARG Detection in Re-Isolated E. coli and Secondary Colonies
4. Discussion
4.1. Effect of Low- and High-Level Contamination by Irrigation
4.1.1. Survival Dynamics of ESBL-E. coli pEK499
4.1.2. Persistence of blaCTX-M-15
4.2. Effect of Contaminated Manure Application
4.2.1. Survival Dynamics of ESBL-E. coli pEK499
4.2.2. Persistence of blaCTX-M-15
4.3. Good Agricultural Practice Recommendations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Primer | Sequence (5’-3’) | Product Size (bp) | Origin |
|---|---|---|---|
| aadA5_F | CTG CGA CAC TGG ACA CAA TC | 502 | This study |
| aadA5_R | CGA GCA AGA GCA AGA ACG AC | ||
| CTX-M-15_F | CCT TAG GTT GAG GCT GGG TG | 760 | This study |
| CTX-M-15_R | TTG TTA GGA AGT GTG CCG CT | ||
| tetA_F | CAG GCA GAG CAA GTA GAG GG | 634 | This study |
| tetA_R | CCT CAA TTT CCT GAC GGG CT | ||
| 27F | AGA GTT TGA TCM TGG CTC AG | 1505 | [65] |
| 1492R | CGG TTA CCT TGT TAC GAC TT |
| 98 °C | Initial Denaturation | 30 sec | |
|---|---|---|---|
| 98 °C | Denaturation | 10 sec | 30 cycles |
| 65 °C | Annealing | 30 sec | |
| 72 °C | Extension | 30 sec | |
| 72 °C | Final Extension | 10 min |
References
- Drissner, D.; Zuercher, U. Microbial safety of fresh fruits and vegetables. In Encyclopedia of Food Safety; Motarjemi, Y., Todd, E., Moy, G., Eds.; Elsevier: Oxford, UK, 2014. [Google Scholar]
- Yeni, F.; Yavaş, S.; Alpas, H.; Soyer, Y. Most common foodborne pathogens and mycotoxins on fresh produce: A review of recent outbreaks. Crit. Rev. Food Sci. Nutr. 2015, 56, 1532–1544. [Google Scholar] [CrossRef]
- Harris, L.; Farber, J.; Beuchat, L.; Parish, M.; Suslow, T.; Garrett, E.; Busta, F. Outbreaks associated with fresh produce: Incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr. Rev. Food Sci. Food Saf. 2003, 2, 78–141. [Google Scholar] [CrossRef]
- Blaak, H.; Van Hoek, A.H.A.M.; Veenman, C.; Van Leeuwen, A.E.D.; Lynch, G.; Van Overbeek, W.M.; Husman, A.M.D.R. Extended spectrum β-lactamase-and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 2014, 168, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Reuland, E.; Al Naiemi, N.; Raadsen, S.A.; Savelkoul, P.H.M.; Kluytmans, J.A.J.W.; Vandenbroucke-Grauls, C.M.J.E. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Poirel, L.; Nordmann, P.; Nüesch-Inderbinen, M.; Hächler, H.; Stephan, R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Agents Chemother. 2016, 60, 2594–2595. [Google Scholar] [CrossRef]
- Awad, R.A.; Abo-Elmagd, E.K.; Raafat, S.A.; Hassan, E.M.; Alrasheedy, Z.E. Prevalence of Vancomycin resistant Enterococci in different food samples. Egypt. J. Med. Microbiol. 2016, 25, 47–55. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Barile, S.; Perozzi, G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011, 6, 275–284. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef]
- Van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. B: Biol. Sci. 2015, 370, 20140087. [Google Scholar] [CrossRef]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal colonization with extended-spectrum beta-lactamase–producing Enterobacteriaceae and risk factors among healthy individuals: A systematic review and metaanalysis. Clin. Infect. Dis. 2016, 63, 310–318. [Google Scholar] [CrossRef]
- Malakar, A.; Snow, D.D.; Ray, C. Irrigation water quality-A contemporary perspective. Water 2019, 11, 1482. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Buergmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Genet. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Agüera, A.; Bayona, J.M.; Cytryn, E.; Fotopoulos, V.; Lambropoulou, D.; Manaia, C.M.; Michael, C.; Revitt, M.; Schröder, P.; et al. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: The knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes-A review. Water Res. 2017, 123, 448–467. [Google Scholar] [CrossRef]
- Aslan, A.; Cole, Z.; Bhattacharya, A.; Oyibo, O. Presence of antibiotic-resistant Escherichia coli in wastewater treatment plant effluents utilized as water reuse for irrigation. Water 2018, 10, 805. [Google Scholar] [CrossRef]
- Fahrenfeld, N.L.; Ema, Y.; Eo’Brien, M.; Pruden, A. Reclaimed water as a reservoir of antibiotic resistance genes: Distribution system and irrigation implications. Front. Microbiol. 2013, 4, 130. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Schwaiger, K.; Harms, K.; Küchenhoff, H.; Kunz, A.; Meyer, K.; Müller, C.; Bauer, J. Sewage sludge and liquid pig manure as possible sources of antibiotic resistant bacteria. Environ. Res. 2010, 110, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Sengelov, G. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 2003, 28, 587–595. [Google Scholar] [CrossRef]
- FAO and WHO Joint FAO/WHO Expert Meeting in Collaboration with OIE on Foodborne Antimicrobial Resistance: Role of the Environment, Crops and Biocides-Meeting Report. Microbiology Risk Assess. 2019. Available online: http://www.fao.org/3/ca6724en/ca6724en.pdf (accessed on 24 September 2020).
- Gou, M.; Hu, H.-W.; Zhang, Y.-J.; Wang, J.-T.; Hayden, H.; Tang, Y.; He, J.-Z. Aerobic composting reduces antibiotic resistance genes in cattle manure and the resistome dissemination in agricultural soils. Sci. Total Environ. 2018, 612, 1300–1310. [Google Scholar] [CrossRef]
- Selvam, A.; Xu, D.; Zhao, Z.; Wong, J.W. Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 2012, 126, 383–390. [Google Scholar] [CrossRef]
- Xie, W.-Y.; Yang, X.-P.; Li, Q.; Wu, L.-H.; Shen, Q.-R.; Zhao, F.-J. Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures. Environ. Pollut. 2016, 219, 182–190. [Google Scholar] [CrossRef]
- Pires, A.F.; Millner, P.D.; Baron, J.; Jay-Russell, M.T. Assessment of current practices of organic farmers regarding biological soil amendments of animal origin in a multi-regional US study. Food Prot. Trends 2018, 38, 347–362. [Google Scholar]
- Global, G.A.P. Inspection Method and Justification Guideline. Integrated Farm Assurance: All Farm Base, Crops Base, Fruit and Vegetables; Version 5.2. Available online: https://www.globalgap.org/uk_en/documents/ (accessed on 24 September 2020).
- Koller, M. Manure for Vegetables: Farm Practice Recommendations for Minimizing Human Pathogenic Bacteria Contamination in Vegetable Production; Forschungsinstitut für bioloigschen Lanbau (FiBL): Frick, Switzerland, 2011. [Google Scholar]
- Gekenidis, M.-T.; Schöner, U.; Von Ah, U.; Schmelcher, M.; Walsh, F.; Drissner, D. Tracing back multidrug-resistant bacteria in fresh herb production: From chive to source through the irrigation water chain. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Erickson, M.C. Internalization of fresh produce by foodborne pathogens. Annu. Rev. Food Sci. Technol. 2012, 3, 283–310. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.B.; Yaron, S.; Matthews, K.R. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 2002, 68, 397–400. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC): Antibiotic/Antimicrobial Resistance-Biggest Threats; c2019. Available online: https://www.cdc.gov/drugresistance/biggest_threats.html (accessed on 20 March 2020).
- Gouvras, G. The European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe 2018; ECDC: Stockholm, Sweden, 2004; Volume 9, p. 2. [Google Scholar] [CrossRef]
- Dahmen, S.; Métayer, V.; Gay, E.; Madec, J.-Y.; Haenni, M. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Veter. Microbiol. 2013, 162, 793–799. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Zurfluh, K.; Peterhans, S.; Hächler, H.; Stephan, R. Assessment of the prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in ready-to-eat salads, fresh-cut fruit, and sprouts from the Swiss market. J. Food Prot. 2015, 78, 1178–1181. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Knapp, C.W.; Christensen, B.T.; McCluskey, S.; Dolfing, J. Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 2016, 6, 21550. [Google Scholar] [CrossRef] [PubMed]
- Gekenidis, M.-T.; Qi, W.; Hummerjohann, J.; Zbinden, R.; Walsh, F.; Drissner, D. Antibiotic-resistant indicator bacteria in irrigation water: High prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS ONE 2018, 13, e0207857. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.U.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The growing genetic and functional diversity of extended spectrum beta-lactamases. BioMed Res. Int. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- Woodford, N.; Carattoli, A.; Karisik, E.; Underwood, A.; Ellington, M.J.; Livermore, D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009, 53, 4472–4482. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Larsson, M.; Robinson, R.C.; Chen, S.L. Direct and convenient measurement of plasmid stability in lab and clinical isolates of E. coli. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tornevi, A.; Bergstedt, O.; Forsberg, B. Precipitation effects on microbial pollution in a river: Lag structures and seasonal effect modification. PLoS ONE 2014, 9, e98546. [Google Scholar] [CrossRef]
- Farnleitner, A.H.; Hocke, L.; Beiwl, C.; Kavka, G.; Zechmeister, T.; Kirschner, A.; Mach, R. Rapid enzymatic detection of Escherichia coli contamination in polluted river water. Lett. Appl. Microbiol. 2001, 33, 246–250. [Google Scholar] [CrossRef]
- Gekenidis, M.-T.; Gossin, D.; Schmelcher, M.; Schöner, U.; Remus-Emsermann, M.N.; Drissner, D. Dynamics of culturable mesophilic bacterial communities of three fresh herbs and their production environment. J. Appl. Microbiol. 2017, 123, 916–932. [Google Scholar] [CrossRef]
- Birkett, C.I.; Ludlam, H.A.; Woodford, N.; Brown, D.F.J.; Brown, N.M.; Roberts, M.T.M.; Milner, N.; Curran, M.D. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum β-lactamases. J. Med. Microbiol. 2007, 56, 52–55. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef]
- Li, J.; Xin, Z.; Zhang, Y.; Chen, J.; Yan, J.; Li, H.; Hu, H. Long-term manure application increased the levels of antibiotics and antibiotic resistance genes in a greenhouse soil. Appl. Soil Ecol. 2017, 121, 193–200. [Google Scholar] [CrossRef]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. mBio 2016, 7, e02227-15. [Google Scholar] [CrossRef]
- Cerqueira, F.; Matamoros, V.; Bayona, J.M.; Berendonk, T.U.; Elsinga, G.; Hornstra, L.M.; Piña, B. Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 2019, 177, 108608. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Bezanson, G.; Delaquis, P.; Bach, S.; McKellar, R.; Topp, E.; Gill, A.; Blais, B.; Gilmour, M. Comparative examination of Escherichia coli O157:H7 survival on romaine lettuce and in soil at two independent experimental sites. J. Food Prot. 2012, 75, 480–487. [Google Scholar] [CrossRef]
- Solomon, E.B.; Pang, H.-J.; Matthews, K.R. Persistence of Escherichia coli O157:H7 on lettuce plants following spray irrigation with contaminated water. J. Food Prot. 2003, 66, 2198–2202. [Google Scholar] [CrossRef] [PubMed]
- Van Der Linden, I.; Cottyn, B.; Uyttendaele, M.; Vlaemynck, G.; Heyndrickx, M.; Maes, M. Survival of enteric pathogens during butterhead lettuce growth: Crop stage, leaf age, and irrigation. Foodborne Pathog. Dis. 2013, 10, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Negreanu, Y.; Pasternak, Z.; Jurkevitch, E.; Cytryn, E. Impact of treated wastewater irrigation on antibiotic resistance in agricultural soils. Environ. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef]
- Dungan, R.S.; McKinney, C.W.; Leytem, A.B. Tracking antibiotic resistance genes in soil irrigated with dairy wastewater. Sci. Total. Environ. 2018, 635, 1477–1483. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.; Pelludat, C.; Gisler, P.; Drissner, D. Conjugation dynamics of self-transmissible and mobilisable plasmids into E. coli O157:H7 on Arabidopsis thaliana rosettes. bioRxiv 2018, 375402. [Google Scholar] [CrossRef]
- Franz, E.; Schijven, J.; Husman, A.M.D.R.; Blaak, H. Meta-regression analysis of commensal and pathogenic Escherichia coli survival in soil and water. Environ. Sci. Technol. 2014, 48, 6763–6771. [Google Scholar] [CrossRef]
- Franz, E.; Semenov, A.; Van Bruggen, A. Modelling the contamination of lettuce with Escherichia coli O157:H7 from manure-amended soil and the effect of intervention strategies. J. Appl. Microbiol. 2008, 105, 1569–1584. [Google Scholar] [CrossRef]
- McKinney, C.W.; Dungan, R.S.; Moore, A.; Leytem, A.B. Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Nõlvak, H.; Truu, M.; Kanger, K.; Tampere, M.; Espenberg, M.; Loit, E.; Raave, H.; Truu, J. Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total. Environ. 2016, 562, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 1–10. [Google Scholar] [CrossRef]
- Fogler, K.W. Effect of Soil Amendments from Antibiotic Treated Cows on Antibiotic Resistant Bacteria and Genes Recovered from the Surfaces of Lettuce and Radishes: Field Study; Virginia Tech: Blacksburg, VA, USA, 2018. [Google Scholar]
- Wang, F.-H.; Qiao, M.; Chen, Z.; Su, J.-Q.; Zhu, Y.-G. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J. Hazard. Mater. 2015, 299, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Urbanowitz, S. Good Agricultural Practices and Good Handling Practices: Water Use in Horticultural Systems. University of Nevada Cooperative Extension. 2013. Available online: https://extension.unr.edu/4h/pub.aspx?PubID=2188 (accessed on 23 September 2020).
- Uyttendaele, M.; Jaykus, L.-A.; Amoah, P.; Chiodini, A.; Cunliffe, D.; Jacxsens, L.; Holvoet, K.; Korsten, L.; Lau, M.; McClure, P.; et al. Microbial hazards in irrigation water: Standards, norms, and testing to manage use of water in fresh produce primary production. Compr. Rev. Food Sci. Food Saf. 2015, 14, 336–356. [Google Scholar] [CrossRef]
- Safe Uses of Agricultural Water. Available online: https://extension.psu.edu/safe-uses-of-agricultural-water (accessed on 19 October 2020).
- Western Growers (WGA). Commodity Specific Food Safety Guidelines for the Production and Harvest of Lettuce and Leafy Greens. Available online: https://www.wga.com/resources/lgma-metrics-californias-new-practices (accessed on 19 October 2020).
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gekenidis, M.-T.; Rigotti, S.; Hummerjohann, J.; Walsh, F.; Drissner, D. Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water. Microorganisms 2020, 8, 1646. https://doi.org/10.3390/microorganisms8111646
Gekenidis M-T, Rigotti S, Hummerjohann J, Walsh F, Drissner D. Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water. Microorganisms. 2020; 8(11):1646. https://doi.org/10.3390/microorganisms8111646
Chicago/Turabian StyleGekenidis, Maria-Theresia, Serena Rigotti, Jörg Hummerjohann, Fiona Walsh, and David Drissner. 2020. "Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water" Microorganisms 8, no. 11: 1646. https://doi.org/10.3390/microorganisms8111646
APA StyleGekenidis, M.-T., Rigotti, S., Hummerjohann, J., Walsh, F., & Drissner, D. (2020). Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water. Microorganisms, 8(11), 1646. https://doi.org/10.3390/microorganisms8111646





