Abstract
Pseudomonas aeruginosa is an opportunistic pathogen displaying high intrinsic antimicrobial resistance and the ability to thrive in different ecological environments. In this study, the ability of P. aeruginosa to develop simultaneous resistance to multiple antibiotics and disinfectants in different natural niches were investigated using strains collected from clinical samples, veterinary samples, and wastewater. The correlation between biocide and antimicrobial resistance was determined by employing principal component analysis. Molecular mechanisms linking biocide and antimicrobial resistance were interrogated by determining gene expression using RT-qPCR and identifying a potential genetic determinant for co- and cross-resistance using whole-genome sequencing. A subpopulation of P. aeruginosa isolates belonging to three sequence types was resistant against the common preservative benzalkonium chloride and showed cross-resistance to fluoroquinolones, cephalosporins, aminoglycosides, and multidrug resistance. Of these, the epidemiological high-risk ST235 clone was the most abundant. The overexpression of the MexAB-OprM drug efflux pump resulting from amino acid mutations in regulators MexR, NalC, or NalD was the major contributing factor for cross-resistance that could be reversed by an efflux pump inhibitor. This is the first comparison of antibiotic-biocide cross-resistance in samples isolated from different ecological niches and serves as a confirmation of laboratory-based studies on biocide adapted isolates. The isolates from wastewater had a higher incidence of multidrug resistance and biocide-antibiotic cross-resistance than those from clinical and veterinary settings.
1. Introduction
Pseudomonas aeruginosa is a ubiquitous bacterium of environmental origin that is responsible for difficult-to-treat nosocomial infections such as wound, urinary tract, and other infections [1,2,3,4]. It is also a major cause of respiratory infections and the main cause of mortality in cystic fibrosis patients [5]. P. aeruginosa is a highly adaptive and robust organism that can thrive in a wide range of environmental niches owing to its large and dynamic genome that provides extraordinary metabolic versatility and genetic plasticity [6]. P. aeruginosa strains display high intrinsic antimicrobial resistance as well as the ability to tolerate a variety of biocides such as antiseptics, disinfectants, and preservatives [6,7,8]. Recently, P. aeruginosa has been classified by the World Health Organization as an organism of the most critical priority in need of new drug development. This is due to the global emergence of multidrug-resistant (MDR) high-risk clones, which are resistant to almost all available antipseudomonal drugs in clinical settings [9,10].
Among the antipseudomonal drugs, fluoroquinolones (FQs) are among the first line of antimicrobials used to treat P. aeruginosa infections [11,12]. FQs are potent, broad-spectrum antimicrobial agents with excellent bioavailability [8] and are characterized by a high degree of persistence in the environment [13]. Several studies have investigated the association between increased consumption of FQs and FQ resistance among P. aeruginosa isolates [14,15], while in vitro experiments have also demonstrated biocide exposure as a driver of antimicrobial resistance [16,17]. Biocides, which include disinfectants, antiseptics, and preservatives, are widely used as a part of infection prevention and control for various applications in both medical and household products [18]. For example, benzalkonium chloride (BKC) is widely used for the disinfection of surfaces and instruments [19] and also as a preservative in non-sterile formulations [20]. Triclosan is used in hand soaps and a large variety of everyday products such as toothpaste, mouthwash, deodorant, shower gel, hand lotion, hand cream, and hand sanitizer [21]. Chlorhexidine is widely used in healthcare for preoperative decontamination and in oral health antiseptics [7]. The use of such biocides unavoidably results in the generation and release of long-lasting residues [19]. The exposure of bacteria to such biocides at subinhibitory concentrations altered the microbial community composition [16,17,22,23] and increased tolerance to biocides as well as resistance to the most critically important antimicrobials used for the treatment of resistant Gram-negative bacteria such as FQs and beta-lactams [16,17,24,25,26]. Such co-selection may result from a number of antimicrobial-resistant (AMR) mechanisms, including drug extrusion by multidrug efflux pumps and mutation of the drug target encoding genes [16,17].
In P. aeruginosa, intrinsic resistance is largely attributed to the expression of multidrug efflux pumps belonging to the Resistance-Nodulation-Division (RND) family [27,28,29]. These pumps are chromosomally encoded membrane proteins forming tripartite complexes composed of an inner membrane transporter protein, a periplasmic adapter protein, and an outer-membrane channel protein [30,31]. Together, these proteins form a highly effective efflux pump able to expel a wide spectrum of structurally unrelated antimicrobial agents from the cell [28]. Genomic analysis has identified structural genes for at least 12 RND type efflux systems, of which four pumps MexAB-OprM, MexCD-OprJ, MexEF-OprN and MexXY-OprM have been confirmed to play a role in MDR [32]. Of these, the MexAB-OprM is the most promiscuous and constitutively expressed multidrug efflux pump in P. aeruginosa and confers basal resistance to a wide range of antimicrobials [28,33]. Exposure to certain substrates or stresses leads to overexpression of the MexAB-OprM efflux pump (predominantly through mutations in regulatory genes such as mexR, nalC, or nalD) [34]. As a result of the promiscuous substrate profile of this efflux pump, the reduced accumulation of many different compounds including antibiotics, dyes, detergents, and disinfectants will be observed, leading to MDR P. aeruginosa [33,35].
In combination with the overexpression of multidrug efflux pumps, repertoires of chromosomal mutations in the quinolone-resistance-determining regions of DNA gyrase and topoisomerase IV-encoding genes gyrA, gyrB, parC, and parE contribute for high-level FQ resistance among P. aeruginosa isolates [36,37,38]. An in vitro experimental study demonstrated that mutations in the gyrA gene combined with overexpression of the MexAB-OprM efflux pump are associated with a high-level resistance to FQ [>16 times increase in FQ minimum inhibitory concentration (MIC) compared to wild type] [24,38].
The highly adaptive and persistent nature of P. aeruginosa [6] coupled with the frequent use of biocides and antimicrobials in healthcare settings exert a further selective pressure on P. aeruginosa to acquire antimicrobial resistance. In particular, wastewater from healthcare sites has been defined as a “hot spot” for the acquisition of antimicrobial resistant determinants [39], as it serves as a reservoir for both antimicrobial residues and bacteria, presenting the perfect environment for the co-selection and development of antimicrobial resistance [19,24,40,41]. Although P. aeruginosa is known to be intrinsically resistant to biocides such as triclosan [42], epidemiologic cut-off values (ECOFF) for biocides are not currently available through the European Committee on Antimicrobial Susceptibility Testing (EUCAST) or Clinical and Laboratory Standards Institute (CLSI), making the determination of biocide resistance difficult.
While the link between increased biocide tolerance and antimicrobial cross-resistance has been experimentally documented in P. aeruginosa [17,24,43], these laboratory-based findings have not yet been observed in diverse natural niches (clinical, veterinary, and wastewater). This provides evidence of a relationship between biocide use and cross-resistance to clinically used antibiotics.
The aim of this study was to assess the correlation between biocide and antimicrobial resistance on a diverse sample set of P. aeruginosa isolates collected from clinical, veterinary, and wastewater sources. Biochemical and in-depth bioinformatic analysis of whole genome sequencing data was employed to investigate the molecular mechanism linking increased biocide resistance to antimicrobial resistance.
2. Materials and Methods
2.1. Source and Identification of P. aeruginosa Isolates
A total of 147 P. aeruginosa isolates were collected: 89 from humans, 20 from companion animals, and 38 from wastewater sources. Clinical isolates were collected from 2012 to 2017 at two hospitals in South Australia, from the USA and from the Netherlands from various clinical infections [44]. Animal isolates were collected in 2013 (Australian collection) from canine otitis, and wastewater isolates were collected from healthcare-generated wastewater in South Australia. Each isolate was sub-cultured on Columbia agar plates and identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) (Bruker Daltonik GmbH, Bremen, Germany). All confirmed isolates were stored at −80 °C for further microbiological and molecular studies.
2.2. Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) for antimicrobials were determined by the broth microdilution technique as described by the EUCAST guidelines [45] and adapted for biocides. Antimicrobial susceptibility was determined according to the 2020 EUCAST (http://www.eucast.org/) ECOFF value. The following classes of antimicrobials and biocides were used: Cephalosporins: ceftazidime, cefepime; Aminoglycosides: gentamicin, tobramycin; Fluoroquinolones: ciprofloxacin, levofloxacin; Carbapenems: imipenem, meropenem; Polymyxins: colistin; Biocides: chlorhexidine digluconate, triclosan, and benzalkonium chloride. Isolates displaying resistance to three or more antimicrobial classes were considered to be MDR [46]. Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were included in each experiment as control strains.
Establishing Biocide Epidemiologic Cut-off Values
The MICs of biocides were determined in triplicate using the broth microdilution technique. ECOFF (with 95% cut-off) values were calculated using the ECOFF finder XL 2010 program (https://clsi.org/meetings/microbiology/ecoffinder/).
2.3. Efflux Pump Inhibition Using a Checkerboard Assay
MICs of biocides and antimicrobials in the presence of the efflux pump inhibitor, phenylalanine arginine β-naphthylamide (PAβN) (Sigma-Aldrich, St. Louis, Missouri, USA) were determined using a checkerboard assay following the steps outlined in a previous study [47]. Only strains that were resistant to FQs (MIC of ciprofloxacin ≥ 1 mg/L and levofloxacin ≥ 2 mg/L) as well as benzalkonium chloride (MIC ≥ 128 mg/L) and triclosan (MIC ≥ 512 mg/L) were subjected to the checkerboard assay in the presence of efflux pump inhibitors. To address the possibility that a high concentration of PAβN permeabilizes the outer membrane of the cell, a nitrocefin hydrolysis assay was performed as described previously [31,48,49]. In brief, a β-lactamase producing P. aeruginosa strain (isolate PA0536 in this study) was treated with PAβN (4 to 128 mg/L) in the presence of nitrocefin (a chromogenic β-lactam) at a final concentration of 32 mg/L. If PAβN causes permeabilization of the outer membrane, nitrocefin will be able to traverse the outer membrane more easily and be exposed to beta-lactamase. This will result in an increased rate of nitrocefin hydrolysis that can be observed as a color change from yellow (≈380 nm) to red (≈490 nm). Polymyxin B, a known outer membrane permeabilizer, was used as a positive control at a final concentration of 128 mg/L. In order to determine the effect of other efflux pumps (not inhibited by PAβN) on the MIC of triclosan, we determined the MIC of triclosan in the presence of carbonyl cyanide-chlorophenylhydrazone (CCCP), which is a proton motive force inhibitor at 12.5 µM using the broth dilution method [50].
2.4. Gene Expression Analysis by Reverse-Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Reverse-transcription quantitative real-time polymerase chain reaction (RT-qPCR) was used to quantify the transcription level of mexA, encoding for the periplasmic adapter protein of the MexAB–OprM drug efflux pump. Isolates shown to be resistant to FQ (MIC of ciprofloxacin ≥ 8 mg/L) and BKC (MIC ≥ 128 mg/L) and showing a ≥ 4-fold reduction in the MIC for both antibacterial agents in the presence of PAβN were selected for RT-qPCR analysis. Overnight cultures of P. aeruginosa were diluted (1:100) and grown at 37 °C in a cation adjusted Muller Hinton broth, up to the mid-log phase (A660 of 0.5) followed by the addition of two times the MIC of ciprofloxacin for 30 min (shock). Total RNA was extracted using a combination of Trizol reagent (Ambion Thermo Fisher Scientific, Carlsbad, CA, USA) and the MN NucleoSpin®RNA (Macherey-Nagel GmbH and Co.KG, Duren Germany) kit following the manufacturer’s instructions. Briefly, 1 mL of cells were pelleted and resuspended in Trizol and chloroform, and further purified using the NucleoSpin®RNA kit. To remove DNA to a completely undetectable level, RNase-free rDNase digestion and further RNA clean-up was performed using the same kit. RNA purity and concentration were determined using a DeNovix DS-11 + Spectrophotometer (DeNovix Inc., Wilmington, DE, USA) and samples stored at −80 °C.
RT-qPCR was performed using a magnetic induction cycler instrument (AdeLab Scientific, Adelaide, Australia) with a KAPA SYBR® FAST One-Step RT-qPCR master mix (2×) kit (Sigma-Aldrich, Sigma-Aldrich, St. Louis, Missouri, USA). A housekeeping gene, rspl, was used to normalize the transcriptional levels of target genes, which were further calibrated against the P. aeruginosa PAO1 control strain. The primer sequences for the target gene (mexA) are: mexA_F 5′-AGACGGTGACCCTGAATACC-3′; mexA_R 5′-GTCGGCCTCGTAGGTGG-3′; rspl_F 5′-CCAACGGTTTCGAGGTTTC-3′; rspl_R 5′-ACCCTGCTTACGGTCTTTGA-3′. Each PCR experiment was carried out in triplicate with the following cycling parameters; a reverse transcription step at 42 °C for 10 min, and reverse transcription inactivation at 95 °C for 3 min, followed by 40 cycles of PCR at 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 15 s for denaturation, annealing and extension, respectively. A no-template control (NTC) and no reverse-transcriptase control (NRT) were included in each experiment. The relative expression of mexA was analyzed using the 2∆∆cq method and normalized against the expression of the PAO1 rspl gene [51].
2.5. DNA Extraction and Whole Genome Sequencing (WGS)
Genomic DNA was extracted using the MN NucleoSpin®Microbial DNA (Machery-Nagel GmbH and Co.KG, Duren, Germany) kit following the manufacturer’s instructions. The quantification of extracted genomic DNA was determined using a DeNovix DS-11 + Spectrophotometer. WGS was conducted at a high-throughput sequencing facility at SA Pathology in South Australia. Sequencing libraries were prepared using the Nextera XT DNA library preparation kit (Illumina Inc., San Diego, Ca, USA), with modifications of the kit’s protocol. Briefly, genomic DNA was fragmented, followed by the amplification of Nextera XT indices (Illumina Inc., San Diego, CA, USA) to the DNA fragments using a low-cycle PCR reaction. The amplicon library was then purified, and normalised manually. Whole-genome sequencing (WGS) was performed on the Illumina NextSeq 550 platform with NextSeq 500/550 Mid-Output kit v2.5 (300 cycles) (Illumina Inc., San Diego, Ca, USA).
2.6. WGS Assembly, Annotation and Analysis
Raw paired-end sequencing reads were assembled and annotated using the TORMES pipeline v.1.1 [52]. Multi-Locus Sequence Type (MLST) assignment was carried out using the mlst software (T. Seemann, https://github.com/tseemann/mlst) and the PubMLST database [53]. A pangenome comparison of all isolates was performed using roary2svg (T. Seemann, https://github.com/sanger-pathogens/Roary/blob/master/contrib/roary2svg/roary2svg.pl). Draft genomes were screened for antimicrobial resistance genes using ResFinder [54], Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) [55], and the comprehensive antibiotic resistance database (CARD) [56]. Genes conferring biocide resistance were identified using the BacMet database (http://bacmet.biomedicine.gu.se) [57]. The P. aeruginosa PAO1 genome was used as a reference strain (accession number NC_002516.2; GI 110645304).
Antimicrobial resistance proteins identified by CARD to be carrying a mutation were used in a protein Basic Local Alignment Search Tool (BLASTp) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to compare regions of similarity between sequences. Protein sequences identified by BLASTp were subsequently used to generate a multi-sequence protein alignment using CLUSTAL O v.1.2.4 [58] to determine the prevalence of the mutation in other proteins.
2.7. Genome Accession Numbers
The draft whole genome sequences for 15 P. aeruginosa isolates has been deposited at GenBank, the accession number of each isolate were as follows: JACVNM000000000 (PA0115), JACVNN000000000 (PA0404), JACVNO000000000 (PA0461), JACVNP000000000 (PA0471), JACVNQ000000000 (PA0507), JACVNR000000000 (PA0508), JACVNS000000000 (PA0532), JACVNT000000000 (PA0536), JACVNU000000000 (PA0545), JACVNV000000000 (PA0550), JACVNW000000000 (PA0555), JACVNX000000000 (PA0570), JACVNY000000000 (PA0571), JACVNZ000000000 (PA0585), JACVOA000000000 (CLN_26).
2.8. Statistical Analysis
Principal component analysis (PCA) was used to cluster isolates based on their antimicrobial resistance patterns, and correlation matrix was used to determine the significant association between biocide and antimicrobial resistance using RStudio, version 3.6.1 [59]. Significant reversion to susceptible MIC in the presence of PAβN was analyzed using the Wilcoxon signed-rank test using IBM SPSS statistical software version 26.0 (SPSS, Armonk, NY, USA). The statistical analysis of mexA expression was performed using student t-test and presented in mean ± standard deviation. A p-value of ≤ 0.05 was considered significant.
3. Results
3.1. P. aeruginosa Isolates from Wastewater Display a High Prevalence for Multidrug Resistance in Contrast to Clinical and Veterinary Isolates
A culture collection of 147 P. aeruginosa isolates was assembled from clinical samples, veterinary samples, and wastewater, and their susceptibility against five different classes of antimicrobials was determined. The highest frequency of resistance was shown to levofloxacin (36.7%), followed by ciprofloxacin (32.7%), ceftazidime (28.6%), cefepime (27.9%), imipenem (20.4%), gentamicin (20.4%), meropenem (16.3%), and tobramycin (12.2%) (Figure 1 and Table S1). None of the isolates were resistant to colistin. Twenty-eight (19%) isolates were resistant to at least three classes of antimicrobials and were defined as MDR. Of these, 53.6% originated from wastewater.
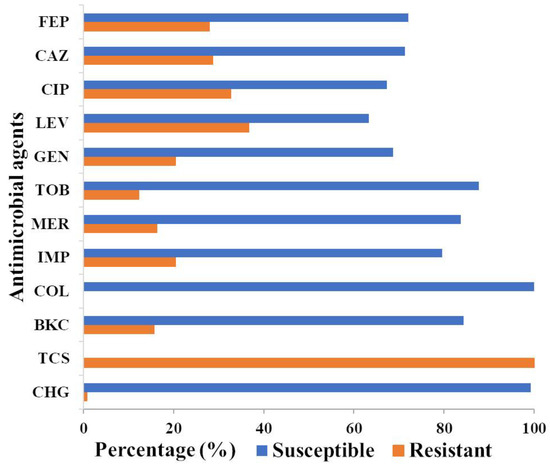
Figure 1.
Proportions of antimicrobial susceptibility versus resistance in P. aeruginosa. Antimicrobial and biocide susceptibility against 147 P. aeruginosa isolates were based on the 2020, European Committee on Antimicrobial Susceptibility Testing (EUCAST) epidemiologic cut-off values (ECOFF) value. The ECOFF values for biocides were as determined in this study (Figure S1), since no ECOFF values were available for biocides against P. aeruginosa. The percentage of susceptible and resistant isolates was indicated in orange and blue, respectively. All 147 isolates of P. aeruginosa were intrinsically resistant to triclosan. FEP, cefepime; CAZ, ceftazidime; CIP, ciprofloxacin; LEV, levofloxacin; GEN, gentamicin; TOB, tobramycin; MER, meropenem; IMP, imipenem; COL, colistin; BKC, benzalkonium chloride; TCS, triclosan, CHG chlorhexidine digluconate.
3.2. P. aeruginosa Has a High Resistance against Common Biocides Used in Clinical Settings
Since antimicrobial resistance is often associated with an increased resistance to biocides, the resistance of P. aeruginosa against common biocides of clinical importance was investigated. There are currently no ECOFF values for biocides against P. aeruginosa; hence, the susceptibility of the 147 P. aeruginosa isolates against the three biocides, BKC, triclosan, and chlorhexidine digluconate, was established to determine the MIC distribution and to establish preliminary ECOFF values as a reference point for measuring resistance (non-wild type). Interestingly, almost all P. aeruginosa isolates were intrinsically resistant to triclosan with an MIC of > 1024 mg/L, only three clinical isolates showed a MIC value ranging from 64 to 256 mg/L (Figure 1, Table S1). The MIC data for chlorhexidine digluconate and BKC exhibit normal distributions, ranging between three and five dilution steps and justified the criteria set by EUCAST for establishing ECOFF values with ECOFF values of 32 and 128 mg/L, respectively (Figure S2). Only one clinical strain displayed resistance to chlorhexidine digluconate, while twenty-three (15.6%) of the isolates had shown an MIC above the wild-type ECOFF value of 128 mg/L to BKC and hence were deemed BKC resistant.
3.3. Phenotypic Correlation between Biocide and Antimicrobial Resistance in P. aeruginosa Isolates
In order to establish if any relationship exists between antimicrobial and biocide resistance, a PCA was performed. Analyses of the MIC values using PCA revealed three clusters of antimicrobial resistance in P. aeruginosa isolates (Figure 2A). The first cluster showed isolates’ resistance to BKC, FQs, and cephalosporins. The second cluster includes isolates resistant to aminoglycosides and carbapenems, while the third cluster exhibits isolates susceptible to colistin and chlorohexidine digluconate (Figure 2A). The MIC values of triclosan were not included in the PCA analysis due to the intrinsic resistance of P. aeruginosa against triclosan (Table S1). To further investigate the dependence between multiple variables at the same time, a correlation matrix was used. The result showed a significant correlation between BKC resistance and the antimicrobial classes FQs, cephalosporins and aminoglycosides (Figure 2B). Since a large number of BKC-tolerant isolates were also resistant to FQs as shown in Figure 2A, further investigation into the underlying mechanisms for cross-resistance between BKC tolerant and ciprofloxacin-resistant subpopulations of P. aeruginosa isolates was carried out.
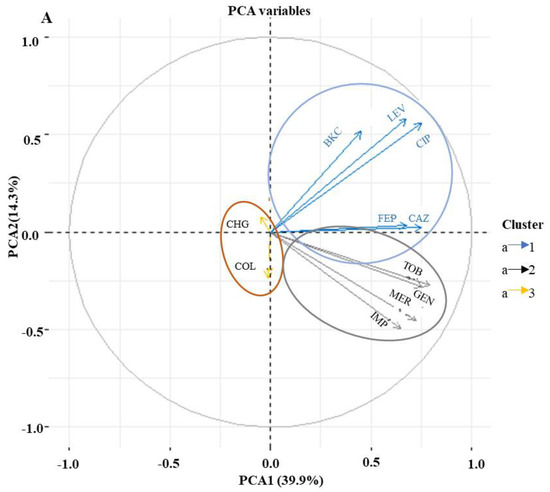
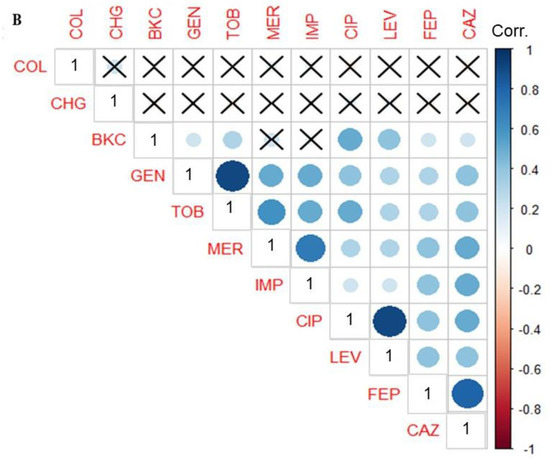
Figure 2.
BKC resistance correlate with antimicrobial resistance. Principal component analysis (PCA) and Pearson correlation coefficient of 147 P. aeruginosa isolates (variables) against five classes of antimicrobials and two biocides based on minimum inhibitory concentration (MIC) value (scores). Colors are used to differentiate isolates with similar clusters of antimicrobial susceptibility patterns (A). The correlation matrix indicates a significant correlation between BKC and antimicrobial classes FQs, cephalosporins, and aminoglycosides. The intensity of the blue circles indicates the correlation coefficient value, and the sizes indicate the significance of correlation (p < 0.05). The bar to the right of the figure indicates the color coding of the correlation value from +1 (strong positive correlation) to −1 (strong negative correlation). Coordinates marked with “×” indicate no significant correlation (p value > 0.05) (B). CAZ, ceftazidime; FEP, cefepime; LEV, levofloxacin; CIP, ciprofloxacin; IMP, imipenem; MER, meropenem; TOB, tobramycin; GEN, gentamicin; COL, colistin; BKC, benzalkonium chloride; CHG chlorhexidine.
3.4. Efflux Pump Inhibition Reveals RND Pump Mediated Biocide and Antimicrobial Resistance
In order to investigate the putative role of drug efflux pumps in the observed antimicrobial and biocide resistance, the effect of the efflux pump inhibitor PAβN on BKC, triclosan, and ciprofloxacin resistance was determined. All BKC-tolerant and 37.5% of the ciprofloxacin-resistant P. aeruginosa isolates were reversed back to sensitive levels when tested in the presence of PAβN (example Figure 3, Table S2). However, PAβN did not modify the MIC of triclosan for any of the isolates tested (Table S2). To explore other potential efflux pumps that may be responsible for triclosan intrinsic resistance but not inhibited by PAβN, we used CCCP. Nonetheless, the reversion of triclosan resistance was not shown upon the addition of CCCP, either (Table S2). To verify that the observed reduction in the MIC of BCK and ciprofloxacin in the presence of PAβN at 32 mg/L was as a direct result of inhibition of the RND-type drug efflux pumps and not due to any non-specific outer membrane permeability effects, the integrity of the outer membrane was determined in the presence of PAβN at 32 mg/L using the nitrocefin hydrolysis rate as an indicator of outer membrane integrity [47,49]. The results clearly indicate no increase in the nitrocefin hydrolysis rate in the presence of PAβN; hence, the reduction in MIC values observed with PAβN was solely due to the inhibition of active efflux (Figure S2).
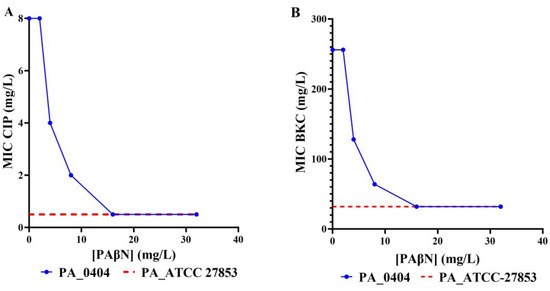
Figure 3.
Reversal of resistance in the presence of an efflux pump inhibitor. Effect of efflux pump inhibitor, (PaβN) in the CIP-resistant and BKC-resistant PA0404 isolate in comparison to a susceptible strain. The MIC values for CIP (A) and BKC (B) of the CIP and BKC-resistant PA0404 isolate were determined in the presence of increasing concentrations of the efflux pump inhibitor PAβN (blue line). For reference, the MIC for the wild-type reference strain—P. aeruginosa ATCC—27853 are indicated, too (red broken line).
3.5. RT-qPCR Revealed MexAB-OprM efflux Pump Overexpression
Due to the effect of the efflux pump inhibitor PAβN on ciprofloxacin and BKC resistance, it was hypothesized that the cross-resistance between these compounds could be mediated by the overexpression of multidrug efflux pumps. MexAB-OprM is the most promiscuous and constitutively expressed efflux pump that has a significant role in natural resistance of P. aeruginosa. To verify the role of MexAB-OprM in cross-resistance, 15 isolates showing an MIC reduction by at least 4-fold in the presence of PAβN were selected for RT-qPCR. Results revealed an overexpression of the MexA pump, showing a 1.896- to 5.911-fold (mean 3.1) difference between the 15 isolates examined and the wild-type PAO1 strain (Table 1, Figure 4). Almost all MexAB-OprM overexpressing isolates are resistant to BKC and showed cross-resistance to FQs resistance, cephalosporins resistance, and MDR subpopulations. The highest level of overexpression (≈6 fold) of MexA was observed for the clinical isolate. Since this clinical isolate was isolated from cystic fibrosis patients, the high-level overexpression is consistent with the role of MexAB-OprM in invasion [60] in addition to its role in antimicrobial resistance.

Table 1.
Phenotypic and genotypic characteristics of the 15 MexAB-OprM efflux pump overexpressing P. aeruginosa isolates, determined by using conventional MICs method, RT-qPCR, and WGS.
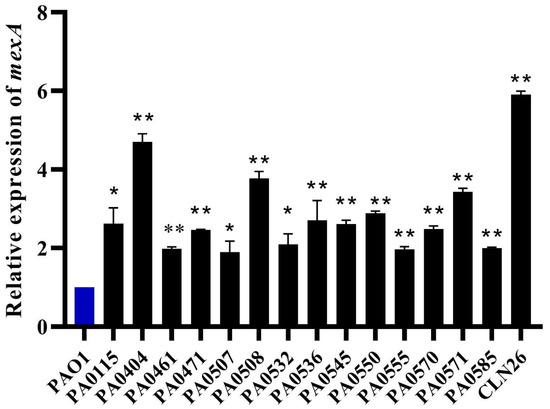
Figure 4.
MexA is overexpressed in BKC-tolerant and ciprofloxacin-resistant isolates. The expression levels of the mexAB-oprM operon, as assessed by RT-qPCR of the mexA gene, reveal the overexpression of mexAB-oprM in BKC-tolerant and CIP-resistant isolates (black bars) as compared to the wild-type PAO1 strain (blue bar) (set at a value of 1, by definition). The results are presented in mean ± standard deviation. Statistical analysis was performed using Student t-test and statistical significance was represented with asterisks (*) as shown in the figure (* p < 0.01; ** p < 0.001).
3.6. WGS Analysis Reveals Multiple Mutations in the Regulators of the mexab-oprM Efflux Pump
WGS analysis was carried out to confirm the molecular mechanism for the link between biocide and FQ resistance in the same 15 P. aeruginosa isolates as mentioned above. Antimicrobial resistance genes were identified using three databases with the CARD identifying not only AMR genes and proteins but also AMR protein variants and their mutations [56]. Further interrogation of the WGS data with blast analysis and multiple sequence alignment revealed multiple amino acid mutations in the regulatory genes of the mexAB-oprM efflux pump (Table 1, Figure 5), which may lead to the observed increased expression of this efflux pump (Figure 4) and subsequent high-level resistance to BKC and FQs. Of these, three non-synonymous mutations were found in NalC proteins, including G71E, S209R, and E153Q, and one mutation (V126E) was found in the MexR regulator. In addition to these mutations, isolate PA0570 had a 13 amino acid truncation at the beginning of the NalC regulator protein. Isolate PA0404, with a fairly high overexpression fold of 4.7, contained an additional T11N mutation in the NalD repressor protein and displayed a novel combination of mutations in the MexR, NalC, and NalD proteins (Table 1).
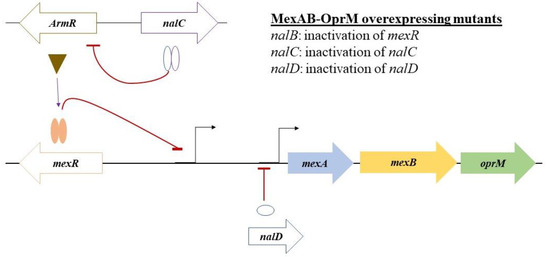
Figure 5.
Schematic representation of the regulator genes involved in the repression of MexAB-OprM efflux pump in P. aeruginosa. The basal expression of the MexAB-OprM efflux pump in wild-type strains is controlled by a local repressor mexR (self-regulated) by binding as a homodimer to the distal promoter of the mexAB-oprM. A second repressor, NalD, directly binds as a monomer to the proximal promoter of the mexAB-oprM Operon. The third repressor, nalC, has an indirect effect on mexAB-oprM expression; it represses the expression of gene armR, the product of which acts as an anti-repressor of mexR. Mutations in these repressors result in a high-level expression of MexAB-OprM, rendering stable, acquired resistance. Mutations in these three repressors have been reported in mexR (nalB mutation), nalD, and nalC mutants, respectively (Table 1).
WGS analysis also revealed that two isolates, PA0508 and PA0532, with MIC values for BKC equal to the ECOFF value, lack the wild-type mexEF-oprJ MDR efflux system and its corresponding regulator genes, mexT and mexS. None of the isolates displayed any mutations in the amino acid sequence of the regulator genes of the mexCD-oprN, mexEF-oprJ, or mexXY-oprM efflux pump systems.
Collectively, the data confirm MexAB-OprM efflux pump overexpression and its role in the cross-resistance of BKC and FQ.
3.7. The Role of Other Resistance Gene Determinants in the Development of High-Level Resistance to FQs and Biocides
Given that 14 sequenced P. aeruginosa isolates displayed high levels of FQ resistance that could not be completely reversed by the efflux pump inhibitor PAβN (Table S2), we investigated the role of other resistance mechanisms using WGS. All 15 of these P. aeruginosa isolates harbored mutations in the DNA gyrase and/or topoisomerase IV proteins (GyrA/B and ParC/E). The main amino acid mutations noted were T83I and D87N in GyrA, S87L and V297I in ParC, and D533E in ParE. A 33 amino acid truncation of the ParC protein was also observed in one isolate (PA0571) as well as a one amino acid deletion in the ParE protein (PA0570). One isolate (CLN26) carried two mutations in the GyrB protein (E468D and H148N) and another carried two mutations in the ParE protein (P438S and L501F) in combination with a T83I mutation in GyrA (Table 1). Although showing no difference to the level of resistance, the ciprofloxacin-modifying crpP gene was also identified in two of the isolates (PA0536 and PA0545). A single amino acid mutation in the GyrA/B or ParC/E (conserved mutations, as observed in these and other P. aeruginosa isolates) is seldom sufficient to confer high-level FQ resistance. However, it is possible that the cumulative effect of these mutations could contribute to the high level of FQ resistance observed.
To screen the presence of resistance genes to BKC and triclosan, the BacMet database was used. All isolates that are resistant to triclosan were shown to carry the triclosan-resistant enoyl-acyl-carrier protein reductase fabV gene encoding for the FabV protein (Table 1).
3.8. The Pangenome Analysis Indicated an Evolutionary Divergence between mexAB-oprM Efflux Pump Overexpressing P. aeruginosa Isolates
To decipher the genetic relationships among MDR P. aeruginosa isolates overexpressing the mexAB-oprM efflux pump system, whole-genome MLST and pangenome analysis was conducted on 15 P. aeruginosa isolates. Three different MLSTs were identified in the study, twelve isolates of which were represented by a single ST, which is the known epidemiological high-risk clone ST235. All of the isolates belonging to ST235 were recovered from wastewater. The remaining P. aeruginosa isolates belonged to ST815 (n = 2), also isolated from wastewater and ST274 (n = 1) isolated from a clinical sample. To further confirm that the dominant ST235 isolates are not diffusion of the same clone as a result of antimicrobial selection pressure, a pangenome analysis was carried out, with the results showing an evolutionary divergence between ST235 isolates (Figure S3).
4. Discussion
Our study on a collection of P. aeruginosa isolated from diverse ecological niches revealed a relatively high proportion (19%) of MDR isolates. This number is much higher than what the Australian Group on Antimicrobial Resistance surveillance found in bacteremic patients (4.3%) [61] and higher than the European Union and European Economic Area surveillance population-weighted mean resistance (12.8%) [62]. The reason for the high proportion of MDR could be the inclusion of isolates from healthcare-generated wastewater which accounted for more than half of the MDR isolates in our study. Multiple factors could contribute to the increased antimicrobial resistance observed in isolates from healthcare wastewater, such as high antimicrobial and disinfectant use in healthcare settings, as well as patients on prolonged courses of antimicrobials who could harbor and shed resistant isolates [19,63].
Most of our isolates originating from wastewater environments demonstrated increased resistance to BKC and cross-resistance for FQ and MDR (55.5% of the total wastewater isolates). This fraction is significantly higher than that observed for isolates from other ecological niches. This difference could be attributed to the difference in the concentration and length of exposure to these agents in these two environments. In the clinical environment, biocides and antibiotics are used at inhibitory concentrations and are less likely to select tolerant clones [64], whereas residues exist at subinhibitory concentrations in the wastewater environment that could provide a selective pressure for the development of resistant subpopulations to thrive and predominate [65].
Studies on laboratory-adapted biocide insensitive P. aeruginosa indicated a correlation with antimicrobial resistance with efflux pump overexpression as the main mechanism of resistance [17,24,66,67,68] and reviewed in [43,69]. These observations have led to the hypothesis that there could be a correlation between biocide use and the development of antimicrobial resistance in clinical or environmental situations, although this has never been translated from laboratory-adapted strains to a confirmation in the natural environment. Our study is the first to provide evidence for the correlation between biocide and antibiotic cross-resistance driven by efflux pump overexpression on isolates from native niches that have “adapted” to their environment. It is also the first study of naturally developed cross-resistance that confirms findings previously only seen in laboratory-based studies. Our work also bestows credence to previous suggestions that healthcare-associated effluent could act as a hotspot for antimicrobial resistance development [39]. The reversal of resistance by the efflux pump inhibitor PAβN, direct measurement of efflux pump expression, and WGS confirmed mutational hyperexpression of the chromosomally encoded MexAB-OprM efflux pump as the main reason for the observed resistance and cross-resistance to antibiotics. The upregulation of MDR efflux pumps provides a fitness advantage that is central to the survival of an organism at low concentrations of a diverse range of xenobiotics such as is found in the wastewater environment [17,24,26,70].
Whole-genome analysis of MLSTs revealed an overrepresentation of a subpopulation of P. aeruginosa wastewater isolates belonging to ST235, which has been described as an MDR gene-enriched international high-risk clone isolated from wastewater [3], human, animal, and other niches [8]. Studies suggest that the global spread of ST235 has been potentially associated with the selective pressure of FQs [9]. Interestingly, we also identified two wastewater isolates that belong to ST815, which appear to be uncommon but are resistant to four major antipseudomonal drugs, including carbapenems. The only record of ST815 comes from a study in Portugal [71]. These isolates also carried the ciprofloxacin-modifying crpP resistance gene (50, 51), which has been shown to marginally increase MICs to FQs through ciprofloxacin phosphorylation [72]. The reduced susceptibility mediated by this enzyme is not to the level of clinical resistance; however, the presence of this enzyme facilitates the selection of mutants that generate quinolone resistance and promote treatment failure [36]. Only a single clinical isolate identified from a cystic fibrosis patient showing cross-resistance between BKC and FQs belongs to ST274. This ST is another epidemic high-risk clone circulating in European countries, mainly Spain, and Australia and has previously been isolated from both cystic and non-cystic fibrosis patients [73,74].
Finally, we found that every P. aeruginosa isolate in this study exhibits an intrinsic resistance to triclosan. Sequencing confirmed the presence of the fabV gene, coding for the triclosan-resistant enoyl-acyl-carrier protein in each of these isolates [42]. Before the US Food and Drug Administration banned the use of triclosan (www.fda.gov) in 2016, triclosan was extensively used in common household products such as antimicrobial soaps and other cleaning products. Given the propensity of triclosan to select for AMR [70], this move would help prevent the further development of triclosan and concomitant AMR. However, due to the acquisition of the fabV gene conferring high-level and stable triclosan resistance, the proportion of triclosan-resistant populations appears to be stable [42]. Biocides are also frequently used in mass-marketed household products, which could lead to the development of resistance and have severe consequences for their clinical usefulness [75,76]
Our study clearly shows that cross-resistance between BKC and FQ, as well as MDR, are driven by hyperexpression of the drug efflux pumps and occurs spontaneously in a natural environment. This should serve as a warning that the unlimited and uncontrolled use of biocides has repercussions as BKC plays a role in the development of resistance against our most useful classes of antimicrobials used to treat resistant Gram-negative infections. It also shows the power of efflux pumps as mechanisms of resistance that continue to evolve, ever expanding their capacity to protect bacterial cells against the very antimicrobials we use for treatment. As such, greater restrain in the use of biocides is needed to preserve the usefulness of these important chemicals and to extend the lifetime of currently used antimicrobials.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1647/s1, Table S1. Minimum inhibitory concentration distribution of antimicrobial agents against P. aeruginosa (n = 147) isolates collected from clinical, veterinary and wastewater, Table S2. Effects of PAβN on the MIC value of P. aeruginosa isolates resistant to an antibiotic (e.g., ciprofloxacin) and biocides: BKC and triclosan. Figure S1: ECOFF value determinations for chlorohexidine digluconate and BKC, Figure S2. Evaluating the outer membrane-permeabilizing activity of PAβN, Figure S3. Visualization of pangenome-analysis of 15 P. aeruginosa isolates.
Author Contributions
Conceptualization, A.A., H.V. and J.T.; experimental, A.A., H.V., A.H.N., L.E.L. and S.A.S., analysis, A.A., H.V., S.A.S. and B.J.H., writing—original draft preparation, A.A.; writing—review and editing, A.A., H.V., S.A.S., M.D.B.L., B.D., L.E.L.; supervision, H.V., M.D.B.L., and S.A.S.; funding acquisition, H.V. All authors have read and reviewed the manuscript for publication.
Funding
Work in our laboratory is funded by the National Health and Medical Research Council (NHMRC, GN1147538), the Medical Research Future Fund (MRFF, GN1152556) and by the University of South Australia. A.A is the recipient of an Australian government research training program for international students (RTPi) scholarship.
Acknowledgments
We thank Sarah Vreugde (Basil Hetzel Institute for Translational Health Research the Queen Elizabeth Hospital and the University of Adelaide) and David Gordon from the Flinders Medical Centre for kindly providing the clinical P. aeruginosa isolates as well as the Australian Centre for Antimicrobial Resistance Ecology (ACARE) for kindly providing the veterinary (dog) P. aeruginosa isolates. We are grateful to Erica Donner, Michael Short, Gianluca Brunetti and Samuel Chol Aleer (Future Industries Institute, University of South Australia) for organizing and conducting wastewater sampling and sample pre-processing.
Conflicts of Interest
The authors declared no conflict of interest.
References
- Snyder, L.; Loman, N.; Faraj, L.; Levi, K.; Weinstock, J.; Boswell, T.; Pallen, M.; Ala’Aldeen, D. Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Eurosurveillance 2013, 18, 42. [Google Scholar] [CrossRef]
- Kumarage, J.; Khonyongwa, K.; Khan, A.; Desai, N.; Hoffman, P.; Taori, S. Transmission of multi-drug resistant Pseudomonas aeruginosa between two flexible ureteroscopes and an outbreak of urinary tract infection: The fragility of endoscope decontamination. J. Hosp. Infect. 2019, 102, 89–94. [Google Scholar] [CrossRef]
- Slekovec, C.; Plantin, J.; Cholley, P.; Thouverez, M.; Talon, D.; Bertrand, X.; Hocquet, D. Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS ONE 2012, 7, e49300. [Google Scholar] [CrossRef]
- Quick, J.; Cumley, N.; Wearn, C.M.; Niebel, M.; Constantinidou, C.; Thomas, C.M.; Pallen, M.J.; Moiemen, N.S.; Bamford, A.; Oppenheim, B. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open 2014, 4, e006278. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef] [PubMed]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tümmler, B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Acquired resistance to chlorhexidine–Is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 2016, 94, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa–Mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 100640. [Google Scholar] [CrossRef] [PubMed]
- Treepong, P.; Kos, V.; Guyeux, C.; Blanc, D.; Bertrand, X.; Valot, B.; Hocquet, D. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cheng, A.C.; Turnidge, J.; Collignon, P.; Looke, D.; Barton, M.; Gottlieb, T. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg. Infect. Dis. 2012, 18, 1453. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Granrot, A.; Ahl, J.; Tham, J.; Resman, F.; Riesbeck, K.; Månsson, F. Antimicrobial combination treatment including ciprofloxacin decreased the mortality rate of Pseudomonas aeruginosa bacteraemia: A retrospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Kümmerer, K.; Al-Ahmad, A.; Mersch-Sundermann, V. Biodegradability of some antibiotics, elimination of the genotoxicity and affection of wastewater bacteria in a simple test. Chemosphere 2000, 40, 701–710. [Google Scholar] [CrossRef]
- Liu, W.-L.; Chang, P.-C.; Chen, Y.-Y.; Lai, C.-C. Impact of fluoroquinolone consumption on resistance of healthcare-associated Pseudomonas aeruginosa. J. Infect. 2012, 64, 335–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Chen, Y.; Jiang, S.; Shen, P.; Lu, X.; Xiao, Y. Association between the rate of fluoroquinolones-resistant gram-negative bacteria and antibiotic consumption from China based on 145 tertiary hospitals data in 2014. BMC Infect. Dis. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Tandukar, M.; Oh, S.; Tezel, U.; Konstantinidis, K.T.; Pavlostathis, S.G. Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ. Sci. Technol. 2013, 47, 9730–9738. [Google Scholar] [CrossRef]
- Kim, M.; Weigand, M.R.; Oh, S.; Hatt, J.K.; Krishnan, R.; Tezel, U.; Pavlostathis, S.G.; Konstantinidis, K.T. Widely used benzalkonium chloride disinfectants can promote antibiotic resistance. Appl. Environ. Microbiol. 2018, 84, e01201-18. [Google Scholar] [CrossRef]
- Health Council of the Netherlands. Resistance due to Disinfectants, Background Report to the Advisory Report Careful Use of Disinfectants. The Hague: Health Council of the Netherlands; Publication No. A16/03E. 2016. Available online: www.healthcouncil.nl. (accessed on 22 October 2020).
- Pereira, B.M.P.; Tagkopoulos, I. Benzalkonium chlorides: Uses, regulatory status, and microbial resistance. Appl. Environ. Microbiol. 2019, 85, e00377-19. [Google Scholar] [CrossRef]
- Marple, B.; Roland, P.; Benninger, M. Safety review of benzalkonium chloride used as a preservative in intranasal solutions: An overview of conflicting data and opinions. Otolaryngol. -Head Neck Surg. 2004, 130, 131–141. [Google Scholar] [CrossRef]
- Weatherly, L.M.; Gosse, J.A. Triclosan exposure, transformation, and human health effects. J. Toxicol. Environ. Health B 2017, 20, 447–469. [Google Scholar] [CrossRef]
- Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Comparing the selective and co-selective effects of different antimicrobials in bacterial communities. Int. J. Antimicrob. Agents 2019, 53, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.R.; Kappell, A.D.; McNamara, P.J. Benzalkonium chloride alters phenotypic and genotypic antibiotic resistance profiles in a source water used for drinking water treatment. Environ. Pollut. 2020, 257, 113472. [Google Scholar] [CrossRef]
- Mc Cay, P.H.; Ocampo-Sosa, A.A.; Fleming, G.T. Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 2010, 156, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.E.; McNamara, P.J. The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 2015, 5, 780. [Google Scholar] [CrossRef]
- Venter, H.; Henningsen, M.L.; Begg, S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017, 61, 1–10. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Poole, K. Pseudomonas aeruginosa: Resistance to the max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar]
- Dreier, J.; Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 660. [Google Scholar] [CrossRef] [PubMed]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Sobel, M.L.; Hocquet, D.; Cao, L.; Plesiat, P.; Poole, K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005, 49, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12. [Google Scholar] [CrossRef]
- Rehman, A.; Patrick, W.M.; Lamont, I.L. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 2019, 68, 1–10. [Google Scholar] [CrossRef]
- Bruchmann, S.; Dötsch, A.; Nouri, B.; Chaberny, I.F.; Häussler, S. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 2013, 57, 1361–1368. [Google Scholar] [CrossRef]
- Rozman, U.; Duh, D.; Cimerman, M.; Turk, S.Š. Hospital wastewater effluent: Hot spot for antibiotic resistant bacteria. J. Watersanitation Hyg. Dev. 2020, 10, 171–178. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef]
- Lien, L.T.Q.; Hoa, N.Q.; Chuc, N.T.K.; Thoa, N.T.M.; Phuc, H.D.; Diwan, V.; Dat, N.T.; Tamhankar, A.J.; Lundborg, C.S. Antibiotics in wastewater of a rural and an urban hospital before and after wastewater treatment, and the relationship with antibiotic use—a one year study from Vietnam. Int. J. Environ. Res. Public Health 2016, 13, 588. [Google Scholar] [CrossRef]
- Zhu, L.; Lin, J.; Ma, J.; Cronan, J.E.; Wang, H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 2010, 54, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Kampf, G. Adaptive microbial response to low-level benzalkonium chloride exposure. J. Hosp. Infect. 2018, 100, e1–e22. [Google Scholar] [CrossRef] [PubMed]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front. Cell. Infect. Microbiol. 2017, 7, 418. [Google Scholar] [CrossRef]
- EUCAST. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 509–515. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mowla, R.; Ji, S.; Guo, L.; Lopes, M.A.D.B.; Jin, C.; Song, D.; Ma, S.; Venter, H. Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 2018, 143, 699–709. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]
- Wang, Y.; Alenzy, R.; Song, D.; Liu, X.; Teng, Y.; Mowla, R.; Ma, Y.; Polyak, S.W.; Venter, H.; Ma, S. Structural optimization of natural product nordihydroguaretic acid to discover novel analogues as AcrB inhibitors. Eur. J. Med. Chem. 2020, 186, 111910. [Google Scholar] [CrossRef]
- Pournaras, S.; Maniati, M.; Spanakis, N.; Ikonomidis, A.; Tassios, P.; Tsakris, A.; Legakis, N.; Maniatis, A. Spread of efflux pump-overexpressing, non-metallo-β-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with bla VIM endemicity. J. Antimicrob. Chemother. 2005, 56, 761–764. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, 566–573. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.J. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Volume 1, p. 409. [Google Scholar]
- Hirakata, Y.; Srikumar, R.; Poole, K.; Gotoh, N.; Suematsu, T.; Kohno, S.; Kamihira, S.; Hancock, R.E.; Speert, D.P. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 2002, 196, 109–118. [Google Scholar] [CrossRef]
- Coombs, G.B.J.; Daley, D.; Collignon, P.; Cooley, L.; Gottlieb, T.; Iredell, J.; Kotsanas, D.; Nimmo, G.; Robson, J.; On behalf of the Australian Group on Antimicrobial Resistance and Australian Commission on Safety and Quality in Health Care. Australian Group on Antimicrobial Resistance Sepsis Outcomes Programs: 2018 Report. Sydney: ACSQHC. Available online: https://www.safetyandquality.gov.au/publications-and-resources/resource-library/agar-sepsis-outcome-programs-2018-report (accessed on 22 October 2020).
- ECDC. Surveillance of Antimicrobial Resistance in Europe 2018. Stockholm: ECDC. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2018 (accessed on 22 October 2020).
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Bondurant, S.W.; Duley, C.M.; Harbell, J.W. Demonstrating the persistent antibacterial efficacy of a hand sanitizer containing benzalkonium chloride on human skin at 1, 2, and 4 h after application. Am. J. Infect. Control 2019, 47, 928–932. [Google Scholar] [CrossRef]
- Kraupner, N.; Ebmeyer, S.; Bengtsson-Palme, J.; Fick, J.; Kristiansson, E.; Flach, C.-F.; Larsson, D.J. Selective concentration for ciprofloxacin resistance in Escherichia coli grown in complex aquatic bacterial biofilms. Environ. Int. 2018, 116, 255–268. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. [Google Scholar] [CrossRef]
- Tetard, A.; Zedet, A.; Girard, C.; Plésiat, P.; Llanes, C. Cinnamaldehyde induces expression of efflux pumps and multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01081-19. [Google Scholar] [CrossRef]
- Kim, M.; Hatt, J.K.; Weigand, M.R.; Krishnan, R.; Pavlostathis, S.G.; Konstantinidis, K.T. Genomic and transcriptomic insights into how bacteria withstand high concentrations of benzalkonium chloride biocides. Appl. Environ. Microbiol. 2018, 84, e00197-18. [Google Scholar] [CrossRef]
- Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Jin, M.; Nguyen, S.H.; Mao, L.; Li, J.; Coin, L.J.; Yuan, Z.; Guo, J. Non-antibiotic antimicrobial triclosan induces multiple antibiotic resistance through genetic mutation. Environ. Int. 2018, 118, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Grosso, F.; Quinteira, S.; Brilhante, M.; Ramos, H.; Peixe, L. Two decades of bla VIM-2-producing Pseudomonas aeruginosa dissemination: An interplay between mobile genetic elements and successful clones. J. Antimicrob. Chemother. 2018, 73, 873–882. [Google Scholar] [CrossRef]
- Chávez-Jacobo, V.M.; Hernández-Ramírez, K.C.; Romo-Rodríguez, P.; Pérez-Gallardo, R.V.; Campos-García, J.; Gutiérrez-Corona, J.F.; García-Merinos, J.P.; Meza-Carmen, V.; Silva-Sánchez, J.; Ramírez-Díaz, M.I. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents Chemother. 2018, 62, e02629-17. [Google Scholar]
- López-Causapé, C.; Sommer, L.M.; Cabot, G.; Rubio, R.; Ocampo-Sosa, A.A.; Johansen, H.K.; Figuerola, J.; Cantón, R.; Kidd, T.J.; Molin, S. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci. Rep. 2017, 7, 5555. [Google Scholar] [CrossRef] [PubMed]
- Ocampo-Sosa, A.A.; Fernández-Martínez, M.; Cabot, G.; Peña, C.; Tubau, F.; Oliver, A.; Martínez-Martínez, L. Draft genome sequence of the quorum-sensing and biofilm-producing Pseudomonas aeruginosa strain Pae221, belonging to the epidemic high-risk clone sequence type 274. Genome Announc. 2015, 3, e01343-14. [Google Scholar] [CrossRef]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.J. Bacterial biocide resistance: A new scourge of the infectious disease world? Arch. Dis. Child. 2019, 104, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).