Metagenomic Insight into Environmentally Challenged Methane-Fed Microbial Communities
Abstract
1. Introduction
2. Materials and Methods
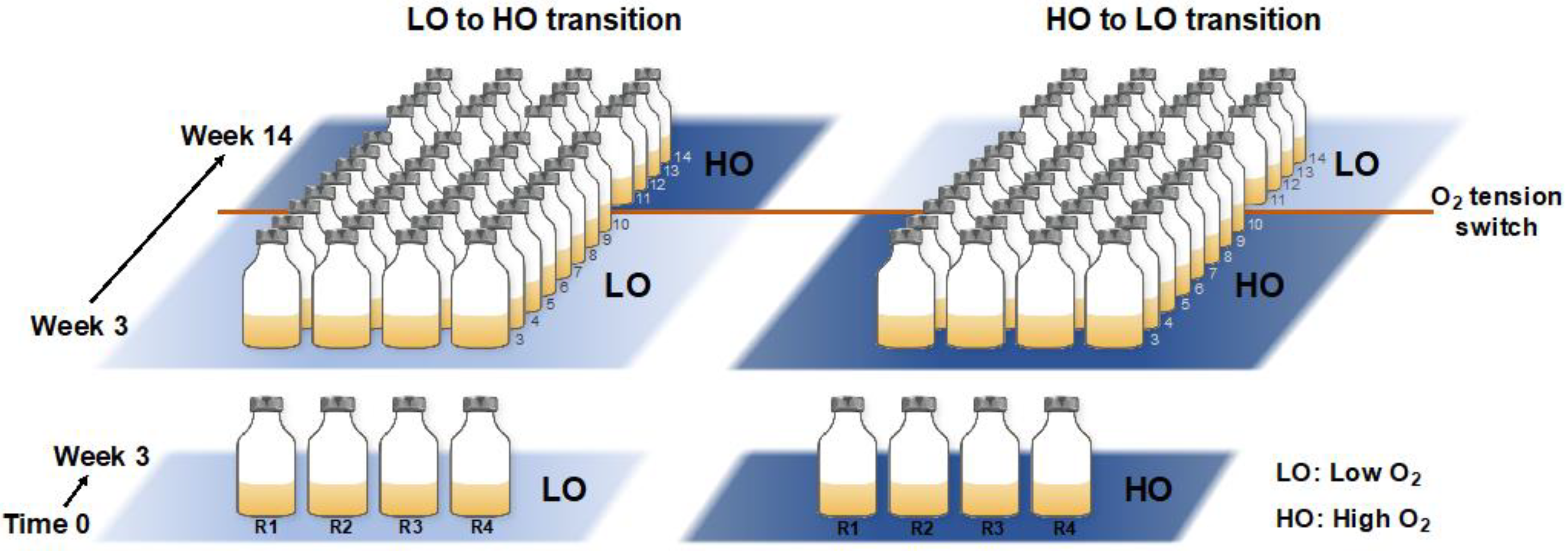
2.1. Experimental Design for Microcosm Manipulation
2.2. DNA and RNA Extraction, Sequencing and Analysis
2.3. Isolation and Identification of Model Organisms
3. Results
3.1. Community Dynamics are Dominated by Four Major Phyla, Firmly Defining the ‘Core Microbiome’ Involved in Methane Consumption
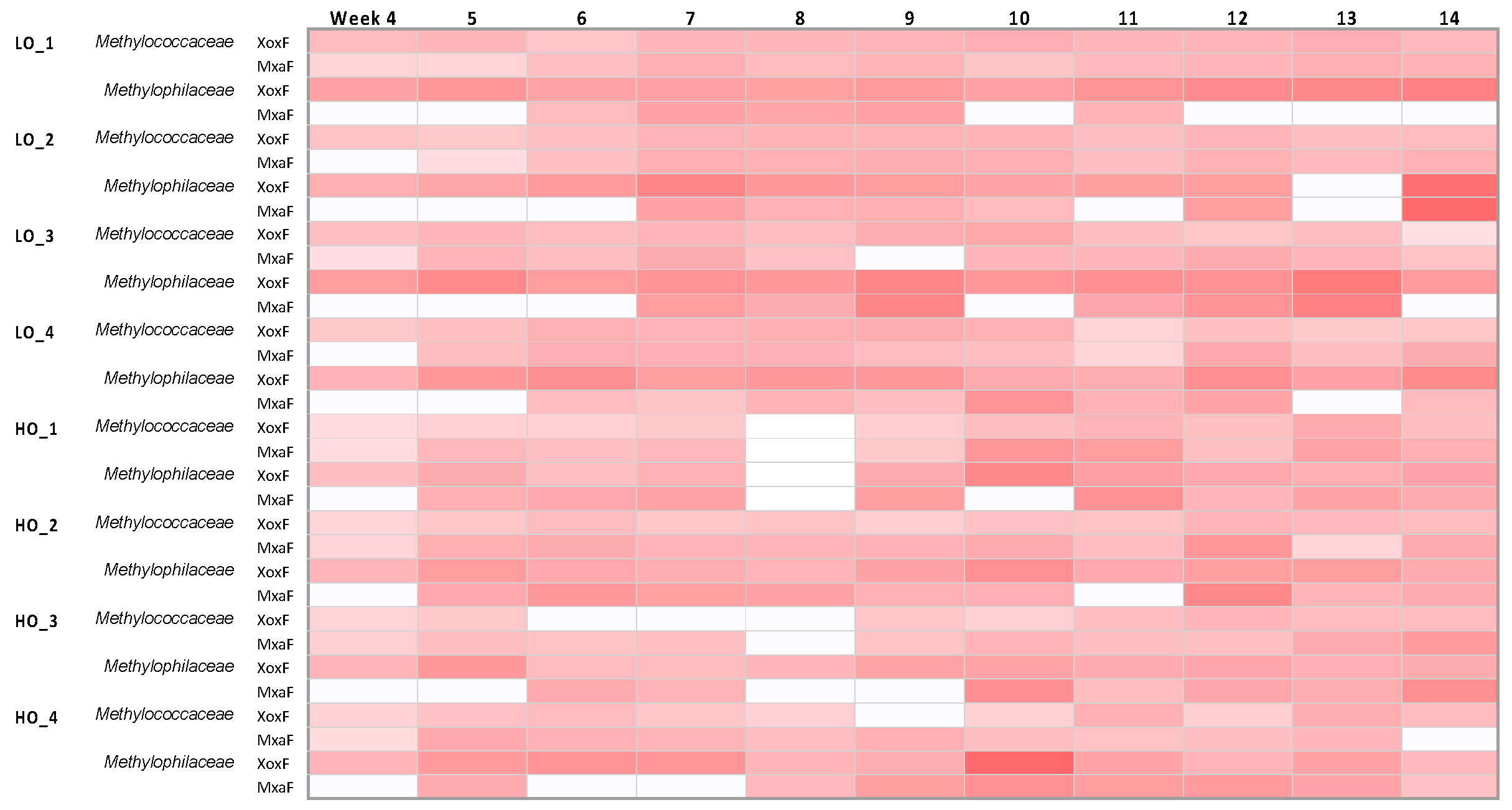
3.2. Community Dynamics within Methylococcaceae and Methylophilaceae Reveal Differential Response to Dioxygen Supply
3.3. Metagenome-Based Genome Assemblies Provide Proxies for Major Players in the Communal Methane Consumption
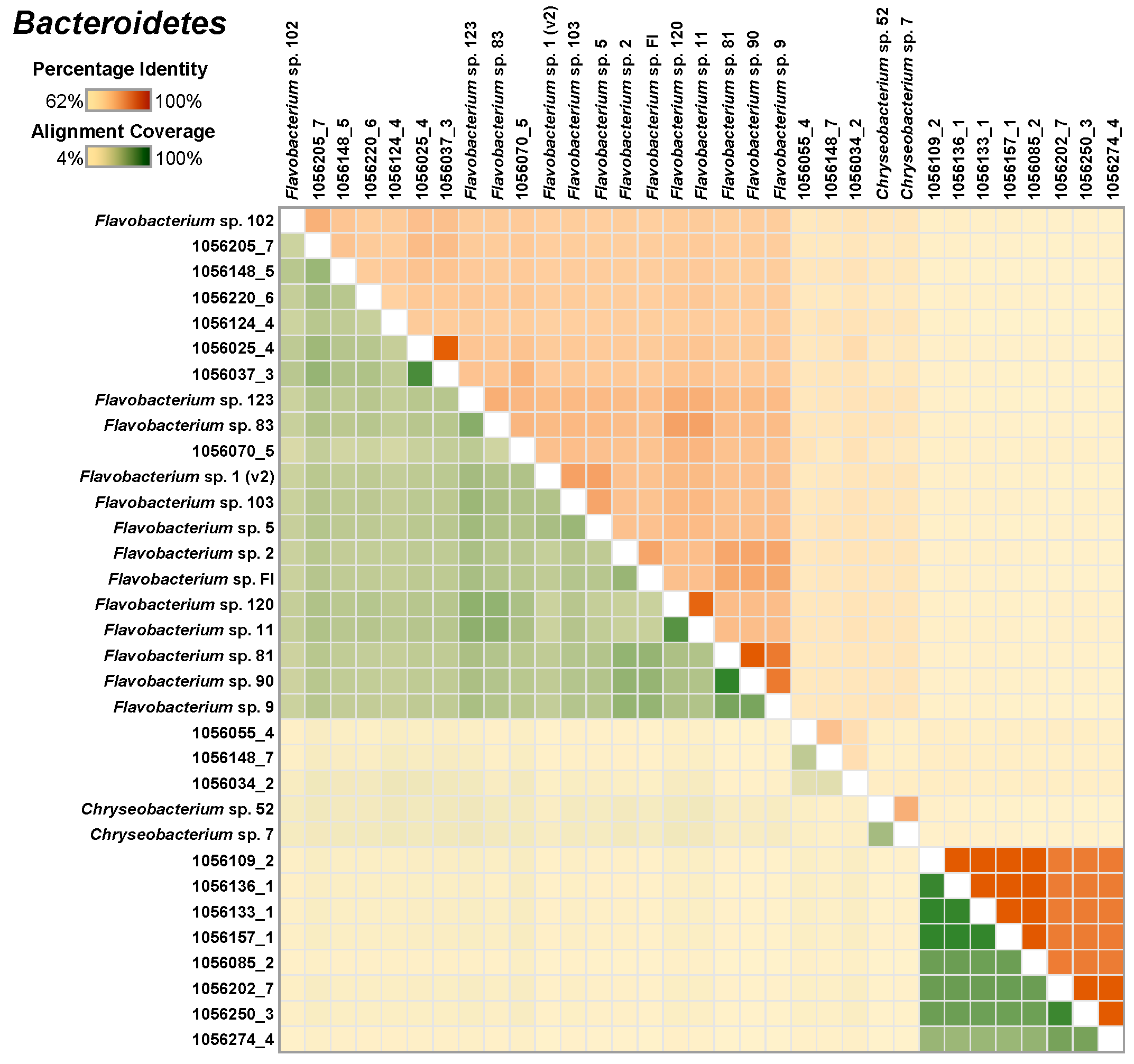
3.4. Organisms Representing the Core Microbiota, Burkholderiales and Bacteropidetes Isolated from Methane-Consuming Microcosms Provide High-Quality Genomic Scaffolds
3.5. Comparative Analysis of Single Genomes and MAGs Identifies the Cultivated Proxies for Community Analysis
3.6. Evidence for Simultaneous Expression of Genes for Alternative Methanol Dehydrogenases in Both Methylococcaceae and Methylophilaceae
3.7. Hypoxia Stress Response Suggests Competition for Dioxygen
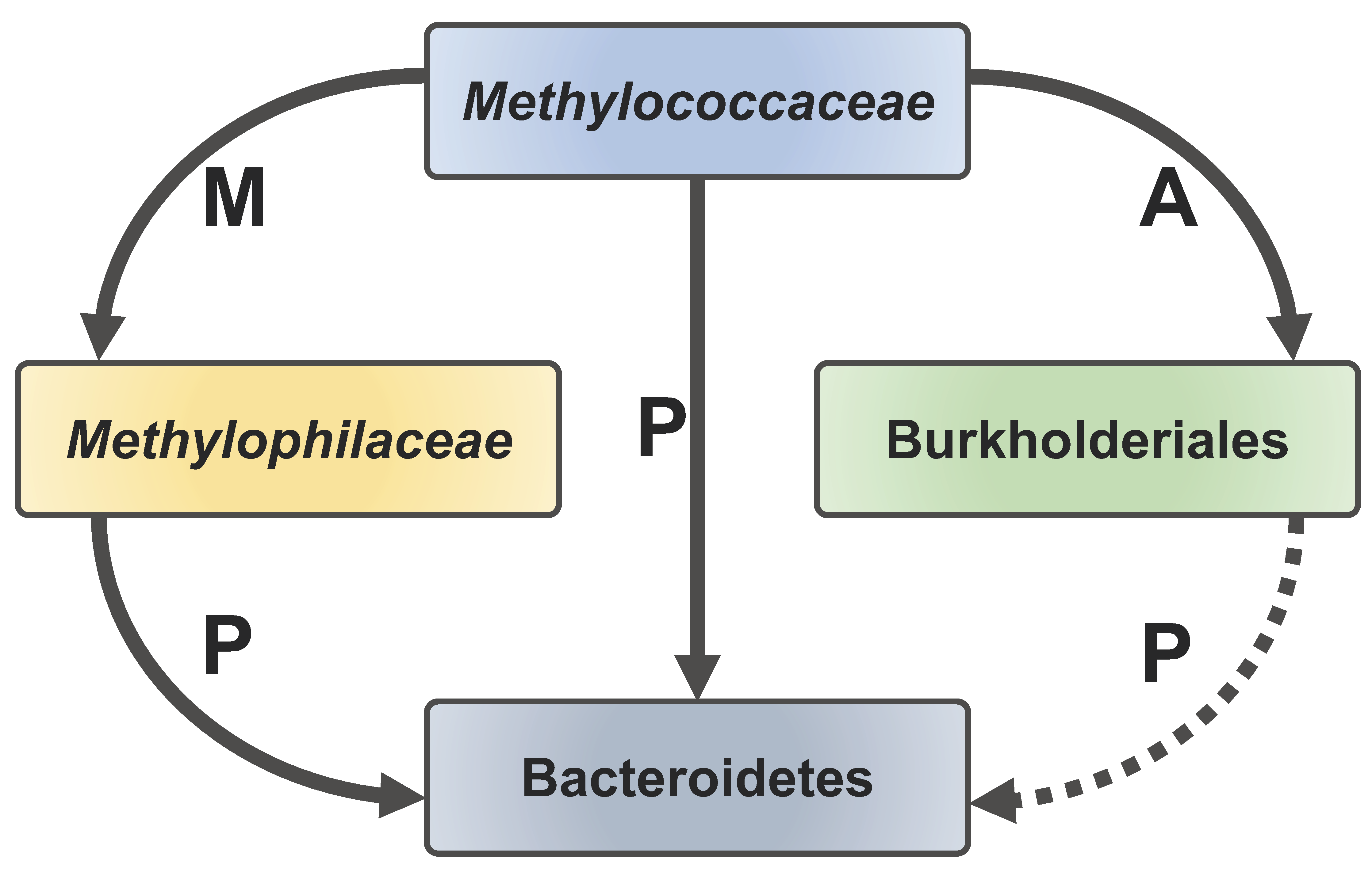
3.8. Metatranscriptome Analysis Uncovers Highly Transcribed Pathways in Burkholderiales and Bacteroidetes, Likely Pinpointing the Mechanisms for the Interspecies Carbon Transfer
3.9. Insights into Metabolisms of Novel Community Members Represented by MAGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sapart, C.J.; Monteil, G.; Prokopiou, M.; van de Wal, R.S.; Kaplan, J.O.; Sperlich, P.; Krumhardt, K.M.; van der Veen, C.; Houweling, S.; Krol, M.C.; et al. Natural and anthropogenic variations in methane sources during the past two millennia. Nature 2012, 490, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.R.; Arora, V.K.; Gauss, M.; Höglund-Isaksson, L.; Parmentier, F.W. Tracing the climate signal: Mitigation of anthropogenic methane emissions can outweigh a large Arctic natural emission increase. Sci. Rep. 2019, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Hmiel, B.; Petrenko, V.V.; Dyonisius, M.N.; Buizert, C.; Smith, A.M.; Place, P.F.; Harth, C.; Beaudette, R.; Hua, Q.; Yang, B.; et al. Preindustrial 14CH4 indicates greater anthropogenic fossil CH4 emissions. Nature 2020, 578, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. In The Prokaryotes, 4th ed.; Rosenberg, E., DeLong, E.F., Thompson, F., Lory, S., Stackebrandt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–285. [Google Scholar]
- Yu, Z.; Chistoserdova, L. Communal metabolism of methane and the rare earth element switch. J. Bacteriol. 2017, 199, e00328-17. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Kalyuzhnaya, M.G. Current trends in methylotrophy. Trends Microbiol. 2018, 26, 703–714. [Google Scholar] [CrossRef]
- Yu, Z.; Zheng, Y.; Huang, J.; Chistoserdova, L. Systems biology meets enzymology: Recent insights into communal metabolism of methane and the role of lanthanides. Curr. Issues Mol. Biol. 2019, 33, 183–196. [Google Scholar] [CrossRef]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R.; et al. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 12, 2544–2558. [Google Scholar] [CrossRef]
- Smith, G.J.; Angle, J.C.; Solden, L.M.; Borton, M.A.; Morin, T.H.; Daly, R.A.; Johnston, M.D.; Stefanik, K.C.; Wolfe, R.; Gil, B.; et al. Members of the genus Methylobacter are inferred to account for the majority of aerobic methane oxidation in oxic soils from a freshwater wetland. MBio 2018, 9, e00815-18. [Google Scholar] [CrossRef]
- Chistoserdova, L. Omics approaches to studying methylotrophs and methylotroph communities. Curr. Issues Mol. Biol. 2017, 24, 119–142. [Google Scholar] [CrossRef]
- Yu, Z.; Pesesky, M.; Zhang, L.; Huang, J.; Winkler, M.; Chistoserdova, L. A complex interplay between nitric oxide, quorum sensing, and the unique secondary metabolite tundrenone constitutes the hypoxia response in Methylobacter. mSystems 2020, 5, e00770-19. [Google Scholar] [CrossRef] [PubMed]
- Danilova, O.V.; Suzina, N.E.; van de Kamp, J.; Svenning, M.M.; Bodrossy, L.; Dedysh, S.N. A new cell morphotype among methane oxidizers: A spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 2016, 10, 2734–2743. [Google Scholar] [CrossRef] [PubMed]
- Oshkin, I.Y.; Miroshnikov, K.K.; Danilova, O.V.; Hakobyan, A.; Liesack, W.; Dedysh, S.N. Thriving in wetlands: Ecophysiology of the spiral-shaped methanotroph Methylospira mobilis as revealed by the complete genome sequence. Microorganisms 2019, 7, 683. [Google Scholar] [CrossRef] [PubMed]
- Oshkin, I.Y.; Beck, D.A.; Lamb, A.E.; Tchesnokova, V.; Benuska, G.; McTaggart, T.L.; Kalyuzhnaya, M.G.; Dedysh, S.N.; Lidstrom, M.E.; Chistoserdova, L. Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J. 2015, 9, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.E.; Beck, D.A.; Lidstrom, M.E.; Chistoserdova, L. Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ 2015, 3, e801. [Google Scholar] [CrossRef]
- Yu, Z.; Beck, D.A.C.; Chistoserdova, L. Natural selection in synthetic communities highlights the roles of Methylococcaceae and Methylophilaceae and suggests differential roles for alternative methanol dehydrogenases in methane consumption. Front. Microbiol. 2017, 8, 2392. [Google Scholar] [CrossRef]
- Krause, S.M.B.; Johnson, T.; Samadhi Karunaratne, Y.; Fu, Y.; Beck, D.A.C.; Chistoserdova, L.; Lidstrom, M.E. Lanthanide-dependent cross-feeding of methane derived carbon is linked by microbial community interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 358–363. [Google Scholar] [CrossRef]
- Yu, Z.; Groom, J.; Zheng, Y.; Chistoserdova, L.; Huang, J. Synthetic methane-consuming communities from a natural lake sediment. mBio 2019, 10, e01072-19. [Google Scholar] [CrossRef]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef]
- Cantor, M.; Nordberg, H.; Smirnova, T.; Hess, M.; Tringe, S.; Dubchak, I. Elviz-exploration of metagenome assemblies with an interactive visualization tool. BMC Bioinform. 2015, 16, 130. [Google Scholar] [CrossRef]
- Kang, D.W.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.; Huntemann, M.; et al. A genomic catalogue of Earth’s microbiomes. Nat. Biotechnol. 2020, in press. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- McTaggart, T.L.; Shapiro, N.; Woyke, T.; Chistoserdova, L. Draft genomes of two strains of Flavobacterium isolated from Lake Washington sediment. Genome Announc. 2015, 3, e01597-14. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Lamb, A.E.; McTaggart, T.L.; Oshkin, I.Y.; Shapiro, N.; Woyke, T.; Chistoserdova, L. Draft genomes of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc. 2015, 3, e00103-15. [Google Scholar] [CrossRef]
- Beck, D.A.; McTaggart, T.L.; Setboonsarng, U.; Vorobev, A.; Kalyuzhnaya, M.G.; Ivanova, N.; Goodwin, L.; Woyke, T.; Lidstrom, M.E.; Chistoserdova, L. The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS ONE 2014, 9, e102458. [Google Scholar] [CrossRef]
- McTaggart, T.L.; Benuska, G.; Shapiro, N.; Woyke, T.; Chistoserdova, L. Draft genomes of five new strains of Methylophilaceae isolated from Lake Washington sediment. Genome Announc. 2015, 3, e01511-14. [Google Scholar] [CrossRef]
- Crossman, L.C.; Chen, H.; Cerdeño-Tárraga, A.M.; Brooks, K.; Quail, M.A.; Pineiro, S.A.; Hobley, L.; Sockett, R.E.; Bentley, S.D.; Parkhill, J.; et al. A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J. 2013, 7, 148–160. [Google Scholar] [CrossRef]
- Smalley, N.E.; Taipale, S.; De Marco, P.; Doronina, N.V.; Kyrpides, N.; Shapiro, N.; Woyke, T.; Kalyuzhnaya, M.G. Functional and genomic diversity of methylotrophic Rhodocyclaceae: Description of Methyloversatilis discipulorum sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 2227–2233. [Google Scholar] [CrossRef]
- McTaggart, T.L.; Shapiro, N.; Woyke, T.; Chistoserdova, L. Draft genome of Pseudomonas sp. 11/12A isolated from Lake Washington sediment. Genome Announc. 2015, 3, e01587-14. [Google Scholar] [CrossRef]
- Rast, P.; Glöckner, I.; Boedeker, C.; Jeske, O.; Wiegand, S.; Reinhardt, R.; Schumann, P.; Rohde, M.; Spring, S.; Glöckner, F.O.; et al. Three novel species with peptidoglycan cell walls form the new genus Lacunisphaera gen. nov. in the family Opitutaceae of the Verrucomicrobial Subdivision 4. Front. Microbiol. 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Dolan, S.K.; Welch, M. The glyoxylate shunt, 60 years on. Ann. Rev. Microbiol. 2018, 72, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.M.; de Jong, A.; Kloosterman, T.G.; Kuipers, O.P.; Saraiva, L.M. The Staphylococcus aureus α-acetolactate synthase ALS confers resistance to nitrosative stress. Front. Microbiol. 2017, 8, 1273. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.J.; Hahnke, R.L.; Huang, S.; Werner, J.; Xing, P.; Barbeyron, T.; Huettel, B.; Stüber, K.; Reinhardt, R.; Harder, J.; et al. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 2013, 79, 6813–6822. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; Cockburn, D.W.; Koropatkin, N.M. The Sus operon: A model system for starch uptake by the human gut Bacteroidetes. Cell. Mol. Life Sci. 2016, 73, 2603–2617. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Fernández-Gómez, B.; Fernàndez-Guerra, A.; Gómez-Consarnau, L.; Sánchez, O.; Coll-Lladó, M.; del Campo, J.; Escudero, L.; Rodríguez-Martínez, R.; Alonso-Sáez, L.; et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. USA 2008, 105, 8724–8729. [Google Scholar] [CrossRef]
- Chen, H.; Brinkac, L.M.; Mishra, P.; Li, N.; Lymperopoulou, D.S.; Dickerson, T.L.; Gordon-Bradley, N.; Williams, H.N.; Badger, J.H. Draft genome sequences for the obligate bacterial predators Bacteriovorax spp. of four phylogenetic clusters. Stand Genom. Sci. 2015, 10, 11. [Google Scholar] [CrossRef]
- Galán, J.E.; Waksman, G. Protein-injection machines in bacteria. Cell 2018, 172, 1306–1318. [Google Scholar] [CrossRef]
- Kumar, R.; Bröms, J.E.; Sjöstedt, A. Exploring the diversity within the genus Francisella—An integrated pan-genome and genome-mining approach. Front. Microbiol. 2020, 11, 1928. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Lührmann, A.; Satoh, A.; Laskowski-Arce, M.A.; Roy, C.R. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 2008, 320, 1651–1654. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.A.; Raftery, D.; Fu, Y.; Bringel, F.; et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef] [PubMed]


| Normalized Reads | Protein Annotation |
|---|---|
| 4406.72 | nitrous-oxide reductase |
| 4361.95 | nitric oxide reductase, NorB subunit |
| 4017.11 | dissimilatory nitrite reductase (NO-forming) |
| 2591.06 | cytochrome c oxidase cbb3-type subunit 1 |
| 2058.85 | nitric oxide reductase, NorC |
| 1840.81 | L-glutamine synthetase |
| 1577.16 | nitrate reductase alpha subunit |
| 1514.15 | cytochrome c oxidase cbb3-type subunit 2 |
| 1511.56 | isocitrate lyase |
| 1464.03 | respiratory nitrate reductase beta subunit |
| 941.95 | nitrogen regulatory protein P-II family |
| 723.80 | nitrate/nitrite transporter NarK |
| 704.82 | Aconitase |
| 685.12 | malate dehydrogenase (NAD) |
| 667.33 | nitrate reductase gamma subunit |
| 642.85 | nitric oxide reductase NorQ |
| 621.24 | type IV pilus assembly protein PilA |
| 617.12 | succinyl-CoA synthetase alpha subunit |
| 615.04 | succinyl-CoA synthetase beta subunit |
| 588.58 | succinate dehydrogenase subunit B |
| 553.90 | acetolactate synthase, small subunit |
| 518.04 | acetolactate synthase, large subunit |
| 517.77 | isocitrate dehydrogenase |
| 425.76 | cytochrome c oxidase cbb3-type subunit 3 |
| 424.15 | 2-isopropylmalate synthase |
| 420.08 | citrate synthase |
| 400.64 | acetyl-coenzyme A synthetase |
| 364.95 | succinate dehydrogenase subunit A |
| 309.10 | isocitrate dehydrogenase |
| 289.19 | dihydrolipoamide dehydrogenase |
| 288.81 | isocitrate lyase |
| 283.16 | glyceraldehyde-3-phosphate dehydrogenase |
| 222.93 | succinate dehydrogenase subunit C |
| 217.08 | Enolase |
| 214.48 | glutamate synthase (NADH) large subunit |
| 198.58 | 6-phosphogluconate dehydratase |
| 196.19 | respiratory nitrate reductase chaperone NarJ |
| 194.83 | pyruvate dehydrogenase E1 component |
| 167.72 | 2-oxoglutarate dehydrogenase E1 component |
| 167.20 | phosphoenolpyruvate synthase |
| 160.71 | NosR/NirI nitrous oxide reductase regulator |
| 160.36 | Transketolase |
| 145.43 | acetyl-CoA carboxylase carboxyltransferase alpha |
| 123.76 | pyruvate dehydrogenase E2 component |
| 116.03 | propionyl-CoA synthetase |
| 114.66 | glutamate synthase (NADH) small subunit |
| 111.75 | fructose-bisphosphate aldolase |
| 110.33 | succinate dehydrogenase subunit D |
| 107.42 | 2-oxoglutarate dehydrogenase E2 component |
| 105.95 | malate synthase |
| Normalized Reads | Protein Annotation |
|---|---|
| 8811.54 | preprotein translocase subunit SecE |
| 2575.34 | protein TonB |
| 2574.02 | outer membrane transport energization protein ExbB |
| 2170.64 | glyceraldehyde 3-phosphate dehydrogenase |
| 1962.49 | preprotein translocase subunit SecG |
| 1556.71 | biopolymer transport protein ExbD |
| 1468.84 | enolase |
| 1430.38 | gliding motility associated protien GldN |
| 1359.97 | pyruvate dehydrogenase E1 component beta subunit |
| 1303.32 | protein involved in gliding motility GldL |
| 1205.79 | fructose-bisphosphate aldolase |
| 1179.97 | citrate synthase |
| 1137.43 | phosphoenolpyruvate carboxykinase |
| 1072.83 | gliding motility-associated lipoprotein GldJ |
| 1058.14 | outer membrane transport energization protein ExbD |
| 1053.74 | protein involved in gliding motility GldK |
| 1028.16 | acetyl-CoA carboxylase carboxyl transferase beta |
| 956.30 | glycine cleavage system H protein |
| 849.13 | cytochrome c oxidase cbb3-type subunit I/II |
| 840.36 | malate dehydrogenase |
| 807.56 | triosephosphate isomerase |
| 717.63 | glycine hydroxymethyltransferase |
| 708.77 | dihydrolipoamide dehydrogenase |
| 682.67 | pyruvate dehydrogenase E1 component alpha subunit |
| 681.09 | succinate dehydrogenase iron-sulfur subunit |
| 673.95 | succinyl-CoA synthetase beta subunit |
| 660.97 | transaldolase |
| 625.88 | glutamine-fructose-6-phosphate transaminase |
| 580.02 | acetyl-CoA carboxylase biotin carboxyl carrier protein |
| 563.70 | acetyl-coenzyme A synthetase |
| 535.31 | starch-binding associating with outer membrane |
| 531.84 | succinyl-CoA synthetase alpha subunit |
| 525.95 | acetyl-CoA carboxylase carboxyl transferase alpha |
| 514.91 | aconitase |
| 509.61 | arabinose-5-phosphate isomerase |
| 508.92 | TonB-linked outer membrane protein, SusC/RagA |
| 507.51 | phosphoglycerate kinase |
| 473.35 | 2-oxoglutarate dehydrogenase E2 component |
| 472.65 | pyruvate dehydrogenase E2 component |
| 439.43 | acetyl-CoA carboxylase, biotin carboxylase subunit |
| 437.56 | 6-phosphofructokinase |
| 427.50 | ribose-phosphate pyrophosphokinase |
| 394.14 | biopolymer transport protein ExbD |
| 377.35 | transketolase |
| 370.57 | starch synthase |
| 363.35 | transketolase |
| 360.88 | 2-oxoglutarate dehydrogenase E1 component |
| 312.13 | 2-oxoglutarate dehydrogenase E2 component |
| 190.55 | Aconitase |
| 190.11 | triosephosphate isomerase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Wang, H.; Yu, Z.; Haroon, F.; Hernández, M.E.; Chistoserdova, L. Metagenomic Insight into Environmentally Challenged Methane-Fed Microbial Communities. Microorganisms 2020, 8, 1614. https://doi.org/10.3390/microorganisms8101614
Zheng Y, Wang H, Yu Z, Haroon F, Hernández ME, Chistoserdova L. Metagenomic Insight into Environmentally Challenged Methane-Fed Microbial Communities. Microorganisms. 2020; 8(10):1614. https://doi.org/10.3390/microorganisms8101614
Chicago/Turabian StyleZheng, Yue, Huan Wang, Zheng Yu, Fauzi Haroon, Maria E. Hernández, and Ludmila Chistoserdova. 2020. "Metagenomic Insight into Environmentally Challenged Methane-Fed Microbial Communities" Microorganisms 8, no. 10: 1614. https://doi.org/10.3390/microorganisms8101614
APA StyleZheng, Y., Wang, H., Yu, Z., Haroon, F., Hernández, M. E., & Chistoserdova, L. (2020). Metagenomic Insight into Environmentally Challenged Methane-Fed Microbial Communities. Microorganisms, 8(10), 1614. https://doi.org/10.3390/microorganisms8101614






