Baltic Group Tick-Borne Encephalitis Virus Phylogeography: Systemic Inconsistency Pattern between Genetic and Geographic Distances
Abstract
1. Introduction
2. Materials and Methods
2.1. Tick Collection
2.2. TBEV Isolation and Sequencing
2.3. Bioinformatical Analysis
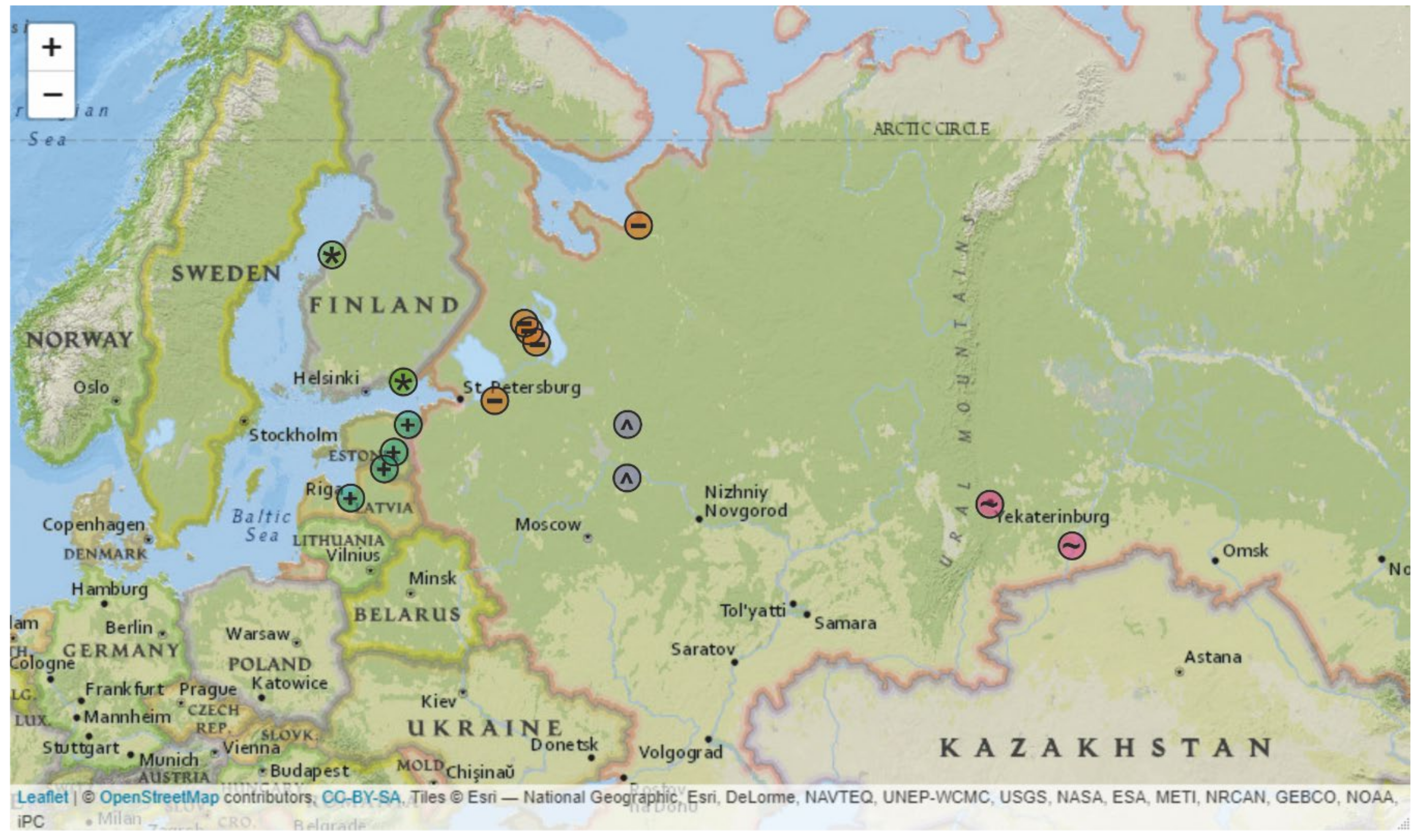
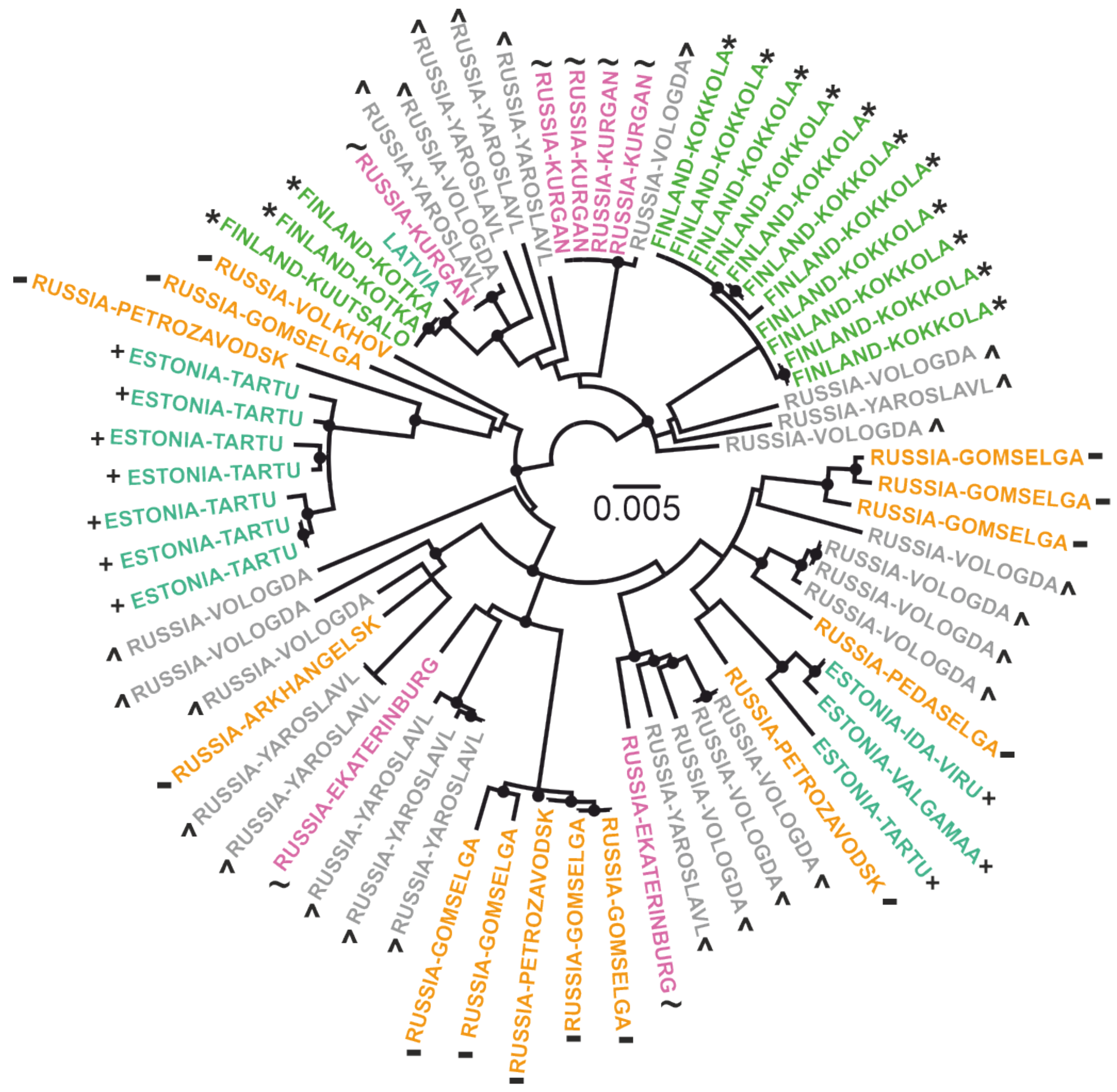
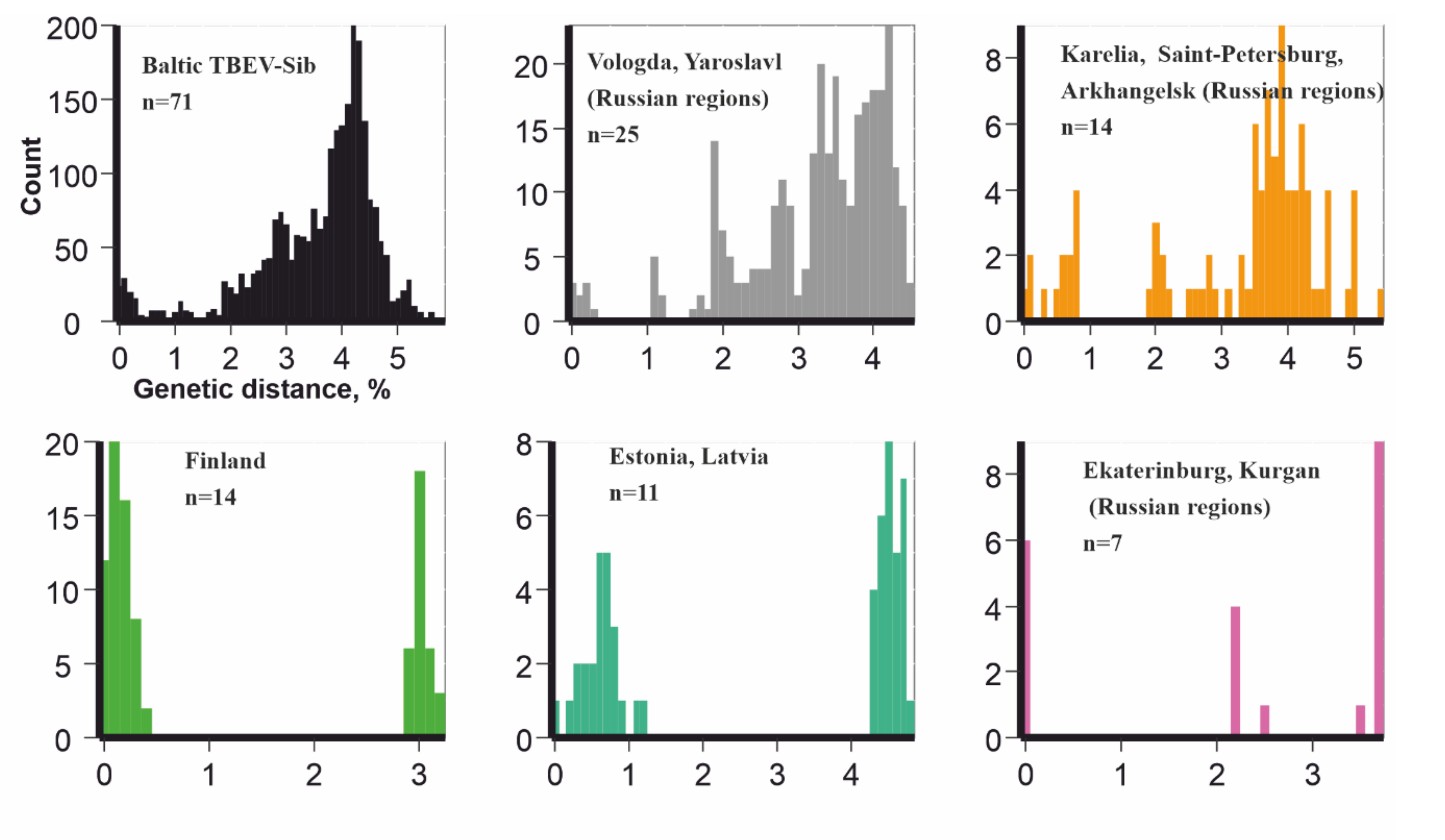
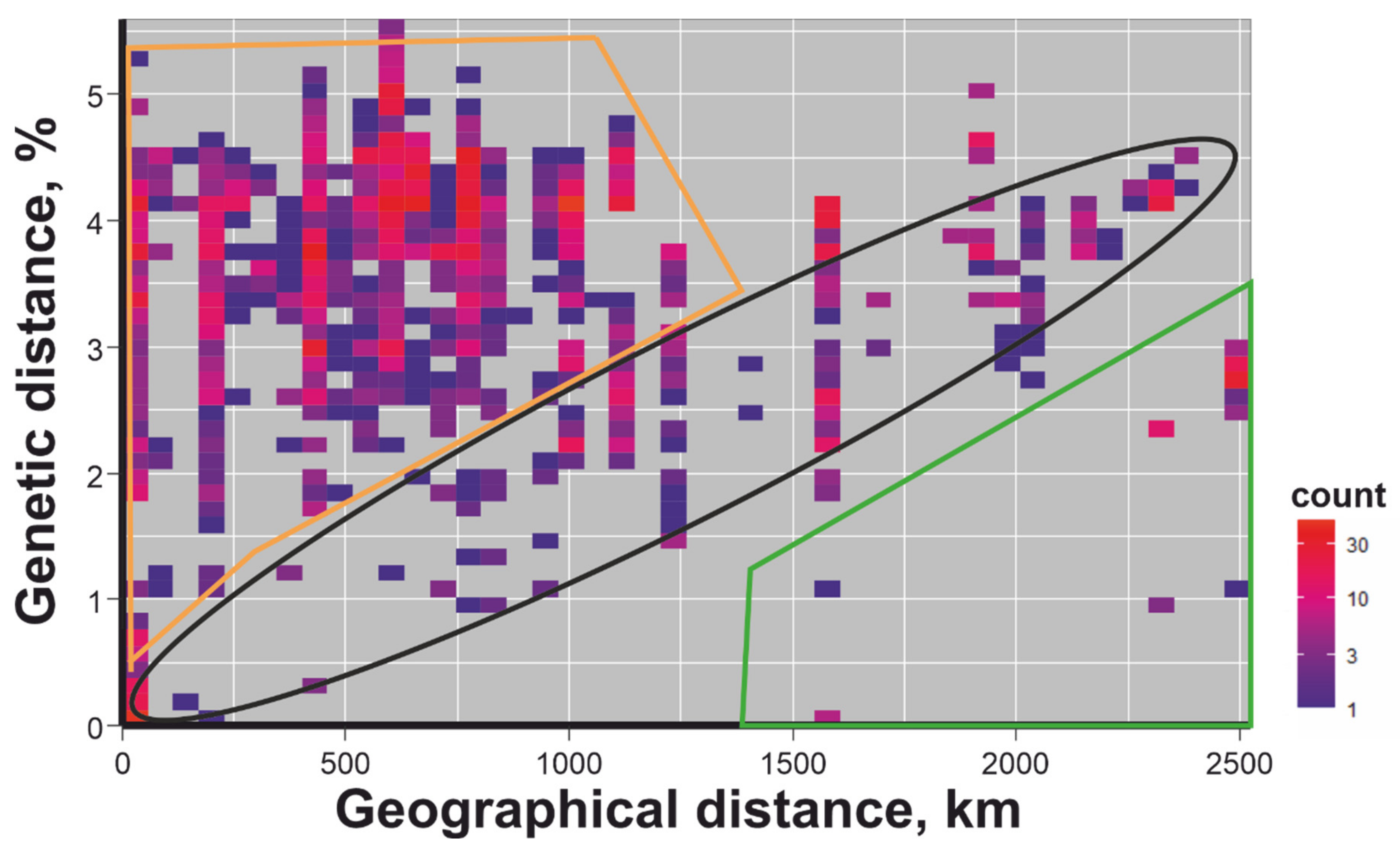
3. Results
4. Discussion
- The known Baltic TBEV-Sib distribution area has borders coinciding with the borders of mixed forest and taiga in Northeastern Europe and the boundaries of Ural mountains;
- I. persulcatus, the main vector for the Siberian TBEV subtype, is spread in the territories from Finland and Estonia in the west to Japan in the east [46];
- Baltic TBEV-Sib was not found in Siberia and the Far-East, where TBEV diversity has been extensively explored;
- Baltic TBEV-Sib is a well-mixed population of viruses with comparable diversity in every region.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Beauté, J.; Spiteri, G.; Warns-Petit, E.; Zeller, H. Tick-borne encephalitis in Europe, 2012 to 2016. Eurosurveillance 2018, 23, 1–9. [Google Scholar] [CrossRef]
- Federal Service for Surveillance on Consumer Rights Protection and HumanWellbeing (Rospotrebnadzor) Statistical Materials. Available online: https://www.rospotrebnadzor.ru/activities/statistical-materials/ (accessed on 13 July 2020).
- Kriz, B.; Hubalek, Z.; Marek, M.; Daniel, M.; Strakova, P.; Betasova, L. Results of the Screening of Tick-Borne Encephalitis Virus Antibodies in Human Sera from Eight Districts Collected Two Decades Apart. Vector-Borne Zoonotic Dis. 2015, 15, 489–493. [Google Scholar] [CrossRef]
- Stjernberg, L.; Holmkvist, K.; Berglund, J. A newly detected tick-borne encephalitis (TBE) focus in south-east Sweden: A follow-up study of TBE virus (TBEV) seroprevalence. Scand. J. Infect. Dis. 2008, 40, 4–10. [Google Scholar] [CrossRef]
- Maikova, G.B.; Chernokhaeva, L.L.; Rogova, Y.V.; Kozlovskaya, L.I.; Kholodilov, I.S.; Romanenko, V.V.; Esyunina, M.S.; Ankudinova, A.A.; Kilyachina, A.S.; Vorovitch, M.F.; et al. Ability of inactivated vaccines based on far-eastern tick-borne encephalitis virus strains to induce humoral immune response in originally seropositive and seronegative recipients. J. Med. Virol. 2019, 91, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; Pullan, S.T.; Lewis, J.; Vipond, R.; Rocchi, M.S.; Baylis, M.; Hewson, R. Tick-Borne Encephalitis Virus, United Kingdom. Emerg. Infect. Dis. 2020, 26, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Agergaard, C.N.; Rosenstierne, M.W.; Bødker, R.; Rasmussen, M.; Andersen, P.H.S.S.; Fomsgaard, A. New tick-borne encephalitis virus hot spot in Northern Zealand, Denmark, October 2019. Eurosurveillance 2019, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tkachev, S.E.E.; Babkin, I.V.V.; Chicherina, G.S.S.; Kozlova, I.V.V.; Verkhozina, M.M.M.; Demina, T.V.V.; Lisak, O.V.V.; Doroshchenko, E.K.K.; Dzhioev, Y.P.P.; Suntsova, O.V.V.; et al. Genetic diversity and geographical distribution of the Siberian subtype of the tick-borne encephalitis virus. Ticks Tick Borne Dis. 2020, 11, 101327. [Google Scholar] [CrossRef]
- Dekker, M.; Laverman, G.D.; de Vries, A.; Reimerink, J.; Geeraedts, F. Emergence of tick-borne encephalitis (TBE) in the Netherlands. Ticks Tick Borne Dis. 2019, 10, 176–179. [Google Scholar] [CrossRef]
- Heinz, F.X.; Stiasny, K.; Holzmann, H.; Kundi, M.; Sixl, W.; Wenk, M.; Kainz, W.; Essl, A.; Kunz, C. Emergence of tick-borne encephalitis in new endemic areas in Austria: 42 years of surveillance. Eurosurveillance 2015, 20, 16–19. [Google Scholar] [CrossRef]
- Ponomareva, E.P.; Mikryukova, T.P.; Gori, A.V.; Kartashov, M.Y.; Protopopova, E.V.; Chausov, E.V.; Konovalova, S.N.; Tupota, N.L.; Gheorghita, S.D.; Burlacu, V.I.; et al. Detection of Far-Eastern subtype of tick-borne encephalitis viral RNA in ticks collected in the Republic of Moldova. J. Vector Borne Dis. 2015, 52, 334–336. [Google Scholar]
- Makenov, M.; Karan, L.; Shashina, N.; Akhmetshina, M.; Zhurenkova, O.; Kholodilov, I.; Karganova, G.; Smirnova, N.; Grigoreva, Y.; Yankovskaya, Y.; et al. First detection of tick-borne encephalitis virus in Ixodes ricinus ticks and their rodent hosts in Moscow, Russia. Ticks Tick Borne Dis. 2019, 10, 101265. [Google Scholar] [CrossRef]
- Botelho-Nevers, E.; Gagneux-Brunon, A.; Velay, A.; Guerbois-Galla, M.; Grard, G.; Bretagne, C.; Mailles, A.; Verhoeven, P.O.; Pozzetto, B.; Gonzalo, S.; et al. Tick-Borne Encephalitis in Auvergne-Rhône-Alpes Region, France, 2017–2018. Emerg. Infect. Dis. 2019, 25, 1944–1948. [Google Scholar] [CrossRef]
- Tokarevich, N.; Tronin, A.; Gnativ, B.; Revich, B.; Blinova, O.; Evengard, B. Impact of air temperature variation on the ixodid ticks habitat and tick-borne encephalitis incidence in the Russian Arctic: The case of the Komi Republic. Int. J. Circumpolar Health 2017, 76, 1–13. [Google Scholar] [CrossRef]
- Wallenhammar, A.; Lindqvist, R.; Asghar, N.; Gunaltay, S.; Fredlund, H.; Davidsson, Å.; Andersson, S.; Överby, A.K.; Johansson, M. Revealing new tick-borne encephalitis virus foci by screening antibodies in sheep milk. Parasit. Vectors 2020, 13, 185. [Google Scholar] [CrossRef]
- Paulsen, K.M.; Neves, C.G.; Granquist, E.G.; Madslien, K.; Stuen, S.; Pedersen, B.N.; Vikse, R.; Rocchi, M.; Laming, E.; Stiasny, K.; et al. Cervids as sentinel-species for tick-borne encephalitis virus in Norway—A serological study. Zoonoses Public Health 2020, 67, 342–351. [Google Scholar] [CrossRef]
- Jaenson, T.G.T.; Värv, K.; Fröjdman, I.; Jääskeläinen, A.; Rundgren, K.; Versteirt, V.; Estrada-Peña, A.; Medlock, J.M.; Golovljova, I. First evidence of established populations of the taiga tick Ixodes persulcatus (Acari: Ixodidae) in Sweden. Parasit. Vectors 2016, 9, 377. [Google Scholar] [CrossRef]
- Brabec, M.; Daniel, M.; Malý, M.; Danielová, V.; Kříž, B.; Kott, I.; Beneš, Č. Analysis of meteorological effects on the incidence of tick-borne encephalitis in the Czech Republic over a thirty-year period. Virol. Res. Rev. 2017, 1, 2–8. [Google Scholar] [CrossRef][Green Version]
- Martello, E.; Mannelli, A.; Ragagli, C.; Ambrogi, C.; Selmi, M.; Ceballos, L.A.; Tomassone, L. Range expansion of Ixodes ricinus to higher altitude, and co-infestation of small rodents with Dermacentor marginatus in the Northern Apennines, Italy. Ticks Tick Borne Dis. 2014, 5, 970–974. [Google Scholar] [CrossRef]
- Ecker, M.; Allison, S.L.; Meixner, T.; Heinz, F.X. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J. Gen. Virol. 1999, 80, 179–185. [Google Scholar] [CrossRef]
- Deviatkin, A.A.; Kholodilov, I.S.; Vakulenko, Y.A.; Karganova, G.G.; Lukashev, A.N. Tick-Borne Encephalitis Virus: An Emerging Ancient Zoonosis? Viruses 2020, 12, 247. [Google Scholar] [CrossRef]
- Jaaskelainen, A.; Tikkakoski, T.; Uzcategui, N.; Alekseev, A.; Vaheri, A.; Vapalahti, O. Siberian Subtype Tickborne Encephalitis Virus, Finland. Emerg. Infect. Dis. 2006, 12, 1568–1571. [Google Scholar] [CrossRef]
- Golovljova, I.; Katargina, O.; Geller, J.; Tallo, T.; Mittženkov, V.; Vene, S.; Nemirov, K.; Kutsenko, A.; Kilosanidze, G.; Vasilenko, V.; et al. Unique signature amino acid substitution in Baltic tick-borne encephalitis virus (TBEV) strains within the Siberian TBEV subtype. Int. J. Med. Microbiol. 2008, 298, 108–120. [Google Scholar] [CrossRef]
- Lundkvist, Å.; Vene, S.; Golovljova, I.; Mavtchoutko, V.; Forsgren, M.; Kalnina, V. Characterization of tick-borne encephalitis virus from Latvia: Evidence for co-circulation of three distinct subtypes. J. Med. Virol. 2001, 65, 730–735. [Google Scholar] [CrossRef]
- Golovljova, I.; Vene, S.; Sjölander, K.B.; Vasilenko, V.; Plyusnin, A.; Lundkvist, Å. Characterization of tick-borne encephalitis virus from Estonia. J. Med. Virol. 2004, 74, 580–588. [Google Scholar] [CrossRef]
- Khasnatinov, M.A.; Ustanikova, K.; Frolova, T.V.; Pogodina, V.V.; Bochkova, N.G.; Levina, L.S.; Slovak, M.; Kazimirova, M.; Labuda, M.; Klempa, B.; et al. Non-hemagglutinating flaviviruses: Molecular mechanisms for the emergence of new strains via adaptation to European ticks. PLoS ONE 2009, 4, 1–11. [Google Scholar] [CrossRef]
- Kovalev, S.Y.; Chernykh, D.N.; Kokorev, V.S.; Snitkovskaya, T.E.; Romanenko, V.V. Origin and distribution of tick-borne encephalitis virus strains of the Siberian subtype in the Middle Urals, the north-west of Russia and the Baltic countries. J. Gen. Virol. 2009, 90, 2884–2892. [Google Scholar] [CrossRef]
- Jaaskelainen, A.E.; Castren, J.; Subbotina, N.; Sironen, T.; Alekseev, A.N.; Vapalahti, O.; Murueva, G.B.; Vaheri, A.; Alitalo, I. Tick-borne encephalitis virus in ticks in Finland, Russian Karelia and Buryatia. J. Gen. Virol. 2010, 91, 2706–2712. [Google Scholar] [CrossRef]
- Pogodina, V.V.; Karan’, L.S.; Koliasnikova, N.M.; Levina, L.S.; Malenko, G.V.; Gamova, E.G.; Lesnikova, M.V.; Kiliachina, A.S.; Esiunina, M.S.; Bochkova, N.G.; et al. Evolution of tick-borne encephalitis and a problem of evolution of its causative agent. Vopr. Virusol. 2007, 52, 16–21. [Google Scholar]
- Available online: https://rpubs.com/andreideviatkin/BalticTBEVdistribution (accessed on 14 October 2020).
- Labuda, M.; Nuttall, P.A. Tick-borne viruses. Parasitology 2004, 129, S221–S245. [Google Scholar] [CrossRef]
- Kovalenko, A.I.; Rubis, L.V.; Ekimova, O.V.; Koloda, N.I. Natural foci infections in Republic of Karelia. EpiNorth 2004, 5, 3Á4. [Google Scholar]
- Lutta, A.S.; Kheysin, E.M.; Shulman, R.E. To the distribution of ticks in Karelia. Quest. Parasitol. Karelia. 1959, 14, 72–83. [Google Scholar]
- Lutta, A.S.; Shul’man, R.E. On the western boundary of Ixodes persulcatus distribution in the territory of Karelian-Finnish ASSR (in Russian). Zool. Zhurnal 1954, 33, 1231–1235. [Google Scholar]
- Bugmyrin, S.V.; Bespyatova, L.A.; Korotkov, Y.S. Long-term dynamics of Ixodes persulcatus (Acari: Ixodidae) abundance in the north–west of its range (Karelia, Russia). Exp. Appl. Acarol. 2019, 77, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Bugmyrin, S.V.; Bespyatova, L.A.; Korotkov, Y.S.; Burenkova, L.A.; Belova, O.A.; Romanova, L.I.; Kozlovskaya, L.I.; Karganova, G.G.; Ieshko, E.P. Distribution of Ixodes ricinus and I. persulcatus ticks in southern Karelia (Russia). Ticks Tick Borne Dis. 2013, 4, 57–62. [Google Scholar] [CrossRef]
- Laaksonen, M.; Sajanti, E.; Sormunen, J.J.; Penttinen, R.; Hänninen, J.; Ruohomäki, K.; Sääksjärvi, I.; Vesterinen, E.J.; Vuorinen, I.; Hytönen, J.; et al. Crowdsourcing-based nationwide tick collection reveals the distribution of Ixodes ricinus and I. persulcatus and associated pathogens in Finland. Emerg. Microbes Infect. 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Sormunen, J.J.; Andersson, T.; Aspi, J.; Bäck, J.; Cederberg, T.; Haavisto, N.; Halonen, H.; Hänninen, J.; Inkinen, J.; Kulha, N.; et al. Monitoring of ticks and tick-borne pathogens through a nationwide research station network in Finland. Ticks Tick Borne Dis. 2020, 11, 101449. [Google Scholar] [CrossRef]
- Filippova, N.A. Ixodid Ticks of the Subfamily Ixodinae. Fauna of the USSR: Arachnoides; Nauka: Leningrad, Russia, 1977; Volume 4. [Google Scholar]
- Kholodilov, I.; Belova, O.; Burenkova, L.; Korotkov, Y.; Romanova, L.; Morozova, L.; Kudriavtsev, V.; Gmyl, L.; Belyaletdinova, I.; Chumakov, A.; et al. Ixodid ticks and tick-borne encephalitis virus prevalence in the South Asian part of Russia (Republic of Tuva). Ticks Tick Borne Dis. 2019, 10, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Schnell, J.K.; Safi, K. Biodiversity Conservation and Phylogenetic Systematics; Pellens, R., Grandcolas, P., Eds.; Topics in Biodiversity and Conservation; Springer International Publishing: Basel, Switzerland, 2016; Volume 14, ISBN 978-3-319-22460-2. [Google Scholar]
- Bessolitsyna, E.A.; Volkov, S.A.; Stolbova, F.S. Dynamics of Ticks’ Infestation with Borrelia Genus Bacteria and Tick-Borne Encephalitis Virus in Kirov Region. Russ. J. Infect. Immun. 2017, 7, 171–180. [Google Scholar] [CrossRef][Green Version]
- Margos, G. Borrelia bavariensis: Vector Switch, Niche Invasion, and Geographical Spread of a Tick-Borne Bacterial Parasite. Front. Ecol. Evol. 2019, 7, 410. [Google Scholar] [CrossRef]
- Lv, J.; De Marco, M.D.M.F.; Goharriz, H.; Phipps, L.P.; McElhinney, L.M.; Hernández-Triana, L.M.; Wu, S.; Lin, X.; Fooks, A.R.; Johnson, N. Detection of tick-borne bacteria and babesia with zoonotic potential in Argas (Carios) vespertilionis (Latreille, 1802) ticks from British bats. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sándor, A.D.; Corduneanu, A.; Péter, Á.; Mihalca, A.D.; Barti, L.; Csősz, I.; Szőke, K.; Hornok, S. Bats and ticks: Host selection and seasonality of bat-specialist ticks in eastern Europe. Parasit. Vectors 2019, 12, 605. [Google Scholar] [CrossRef]
- Jaunbauere, G.; Salmane, I.; Spungis, V. Occurrence of bat ectoparasites in Latvia. Latv. Entomol. 2008, 45, 38–42. [Google Scholar]
- Klaus, C.; Gethmann, J.; Hoffmann, B.; Ziegler, U.; Heller, M.; Beer, M. Tick infestation in birds and prevalence of pathogens in ticks collected from different places in Germany. Parasitol. Res. 2016, 115, 2729–2740. [Google Scholar] [CrossRef]
- Lommano, E.; Dvořák, C.; Vallotton, L.; Jenni, L.; Gern, L. Tick-borne pathogens in ticks collected from breeding and migratory birds in Switzerland. Ticks Tick Borne Dis. 2014, 5, 871–882. [Google Scholar] [CrossRef]
- Mikryukova, T.P.; Moskvitina, N.S.; Kononova, Y.V.; Korobitsyn, I.G.; Kartashov, M.Y.; Tyuten’kov, O.Y.; Protopopova, E.V.; Romanenko, V.N.; Chausov, E.V.; Gashkov, S.I.; et al. Surveillance of tick-borne encephalitis virus in wild birds and ticks in Tomsk city and its suburbs (Western Siberia). Ticks Tick Borne Dis. 2014, 5, 145–151. [Google Scholar] [CrossRef]
- Hasle, G. Transport of ixodid ticks and tick-borne pathogens by migratory birds. Front. Cell. Infect. Microbiol. 2013, 4, 1–6. [Google Scholar] [CrossRef]
- Michelitsch, A.; Wernike, K.; Klaus, C.; Dobler, G.; Beer, M. Exploring the reservoir hosts of tick-borne encephalitis virus. Viruses 2019, 11, 1–17. [Google Scholar] [CrossRef]
- Waldenström, J.; Lundkvist, A.; Falk, K.I.; Garpmo, U.; Bergström, S.; Lindegren, G.; Sjöstedt, A.; Mejlon, H.; Fransson, T.; Haemig, P.D.; et al. Migrating birds and tickborne encephalitis virus. Emerg. Infect. Dis. 2007, 13, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Havlik, O.; Kolman, J. The demonstration of antibodies against the virus of the tick-borne encephalitis in certain bats. J. Hyg. Epidemiol. Microbiol. Immunol. 1957, 1, 231. [Google Scholar]
- Drobishchenko, N.I.; L’vov, D.K.; Ukbaeva, T.D.; Kiriushchenko, T.V.; Karimov, S.K. Experimental corroboration of the persistence of the tick-borne encephalitis virus in bats in wintertime. Med. Parazitol. 1978, 47, 81–82. [Google Scholar]
- Nosek, J.; Gresikova, M.; Rehacek, J. Persistence of Tick-Borne Encephalitis Virus in Hibernating Bats. Acta Virol. 1961, 5, 112–116. [Google Scholar]
- Fagre, A.C.; Kading, R.C. Can bats serve as reservoirs for Arboviruses? Viruses 2019, 11, 215. [Google Scholar] [CrossRef]
- Geller, J.; Nazarova, L.; Katargina, O.; Leivits, A.; Järvekülg, L.; Golovljova, I. Tick-Borne Pathogens in Ticks Feeding on Migratory Passerines in Western Part of Estonia. Vector-Borne Zoonotic Dis. 2013, 13, 443–448. [Google Scholar] [CrossRef]
- Kazarina, A.; Japiņa, K.; Keišs, O.; Salmane, I.; Bandere, D.; Capligina, V.; Ranka, R. Detection of tick-borne encephalitis virus in I. ricinus ticks collected from autumn migratory birds in Latvia. Ticks Tick Borne Dis. 2015, 6, 178–180. [Google Scholar] [CrossRef]
- Csank, T.; Korytár, Ľ.; Pošiváková, T.; Bakonyi, T.; Pistl, J.; Csanády, A. Surveillance on antibodies against West Nile virus, Usutu virus, tick-borne encephalitis virus and Tribeč virus in wild birds in Drienovská wetland, Slovakia. Biologia 2019, 74, 813–820. [Google Scholar] [CrossRef]
- Irwin, D.E.; Irwin, J.H.; Price, T.D. Ring species as bridges between microevolution and speciation. Genetica 2001, 112–113, 223–243. [Google Scholar] [CrossRef]
- Kvist, L.; Martens, J.; Higuchi, H.; Nazarenko, A.A.; Valchuk, O.P.; Orell, M. Evolution and genetic structure of the great tit (Parus major) complex. Proc. R. Soc. B Biol. Sci. 2003, 270, 1447–1454. [Google Scholar] [CrossRef]
- Alcaide, M.; Scordato, E.S.C.; Price, T.D.; Irwin, D.E. Genomic divergence in a ring species complex. Nature 2014, 511, 83–85. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deviatkin, A.A.; Kholodilov, I.S.; Belova, O.A.; Bugmyrin, S.V.; Bespyatova, L.A.; Ivannikova, A.Y.; Vakulenko, Y.A.; Lukashev, A.N.; Karganova, G.G. Baltic Group Tick-Borne Encephalitis Virus Phylogeography: Systemic Inconsistency Pattern between Genetic and Geographic Distances. Microorganisms 2020, 8, 1589. https://doi.org/10.3390/microorganisms8101589
Deviatkin AA, Kholodilov IS, Belova OA, Bugmyrin SV, Bespyatova LA, Ivannikova AY, Vakulenko YA, Lukashev AN, Karganova GG. Baltic Group Tick-Borne Encephalitis Virus Phylogeography: Systemic Inconsistency Pattern between Genetic and Geographic Distances. Microorganisms. 2020; 8(10):1589. https://doi.org/10.3390/microorganisms8101589
Chicago/Turabian StyleDeviatkin, Andrei A., Ivan S. Kholodilov, Oxana A. Belova, Sergey V. Bugmyrin, Lubov A. Bespyatova, Anna Y. Ivannikova, Yulia A. Vakulenko, Alexander N. Lukashev, and Galina G. Karganova. 2020. "Baltic Group Tick-Borne Encephalitis Virus Phylogeography: Systemic Inconsistency Pattern between Genetic and Geographic Distances" Microorganisms 8, no. 10: 1589. https://doi.org/10.3390/microorganisms8101589
APA StyleDeviatkin, A. A., Kholodilov, I. S., Belova, O. A., Bugmyrin, S. V., Bespyatova, L. A., Ivannikova, A. Y., Vakulenko, Y. A., Lukashev, A. N., & Karganova, G. G. (2020). Baltic Group Tick-Borne Encephalitis Virus Phylogeography: Systemic Inconsistency Pattern between Genetic and Geographic Distances. Microorganisms, 8(10), 1589. https://doi.org/10.3390/microorganisms8101589





