Study on the Identification Methods for Effective Microorganisms in Commercially Available Organic Agriculture Materials
Abstract
1. Introduction
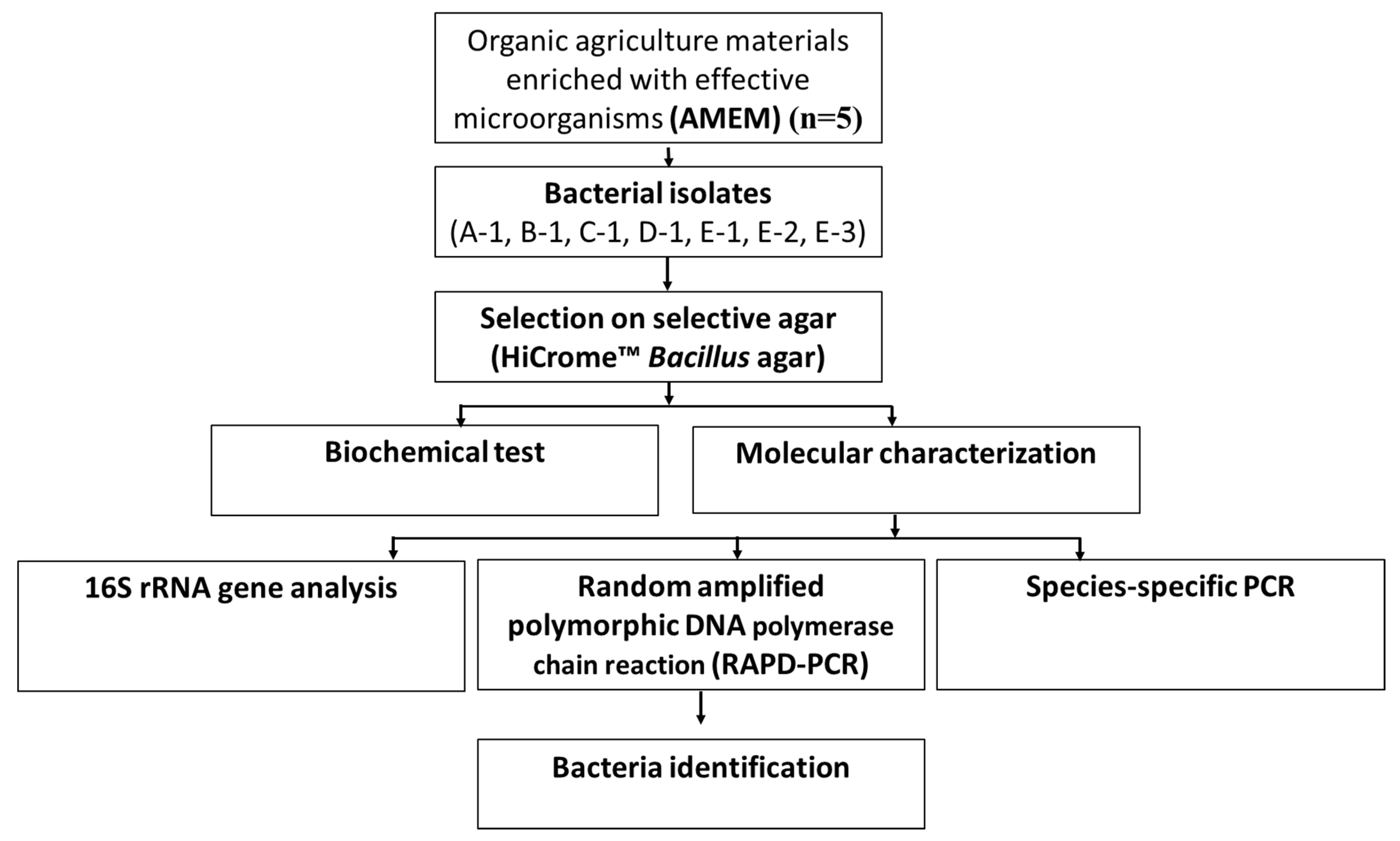
2. Materials and Methods
2.1. Microorganisms and AMEM Products
2.2. Isolation of Bacteria from AMEM
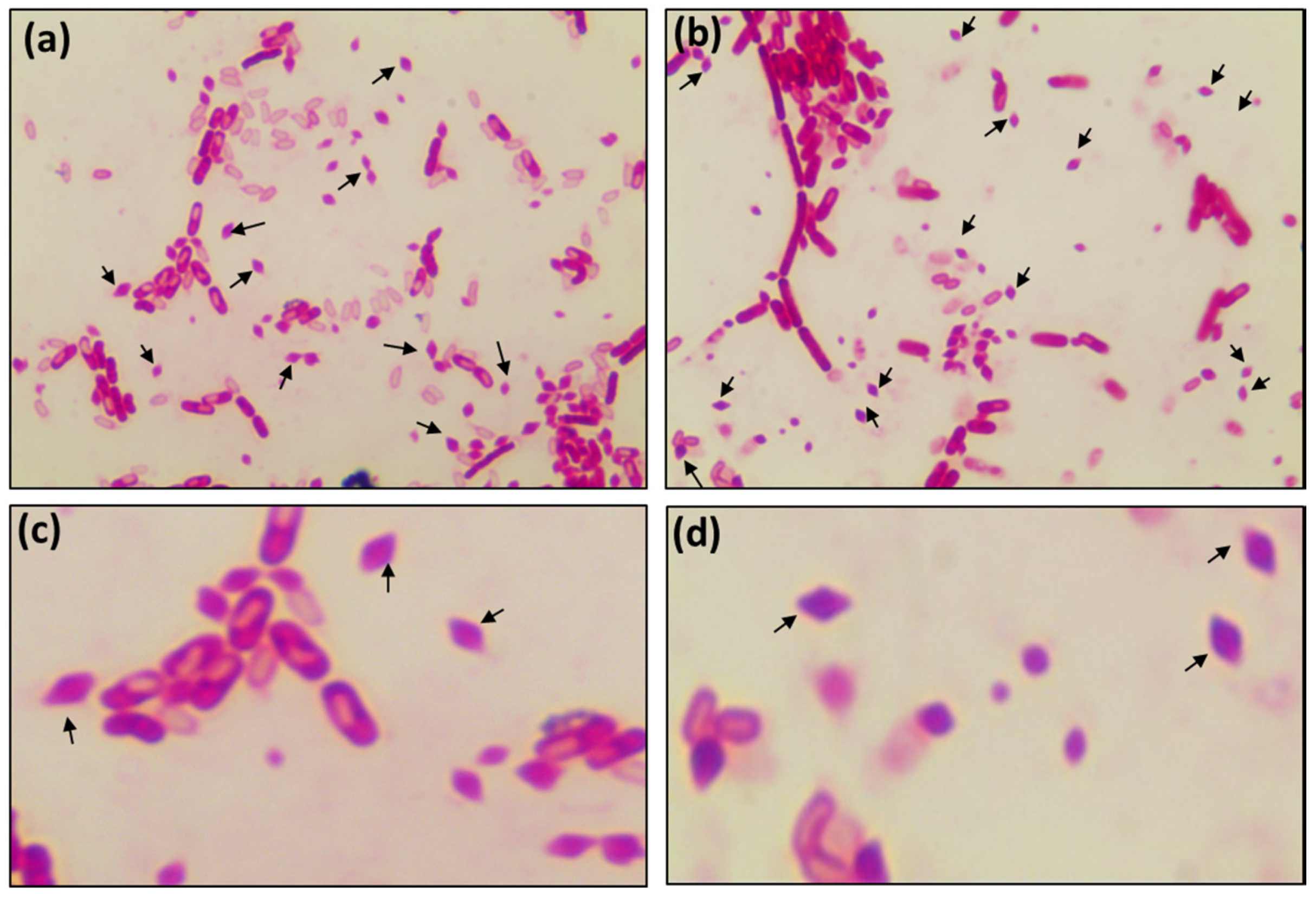
2.3. Morphological Characteristics of AMEM Isolates
2.4. Differential Properties and Biochemical Identification
2.5. 16S rRNA Gene Sequencing Analysis
2.6. RAPD-PCR
2.7. Species-Specific PCR
2.8. Crystalline Protein Staining
3. Results and Discussion
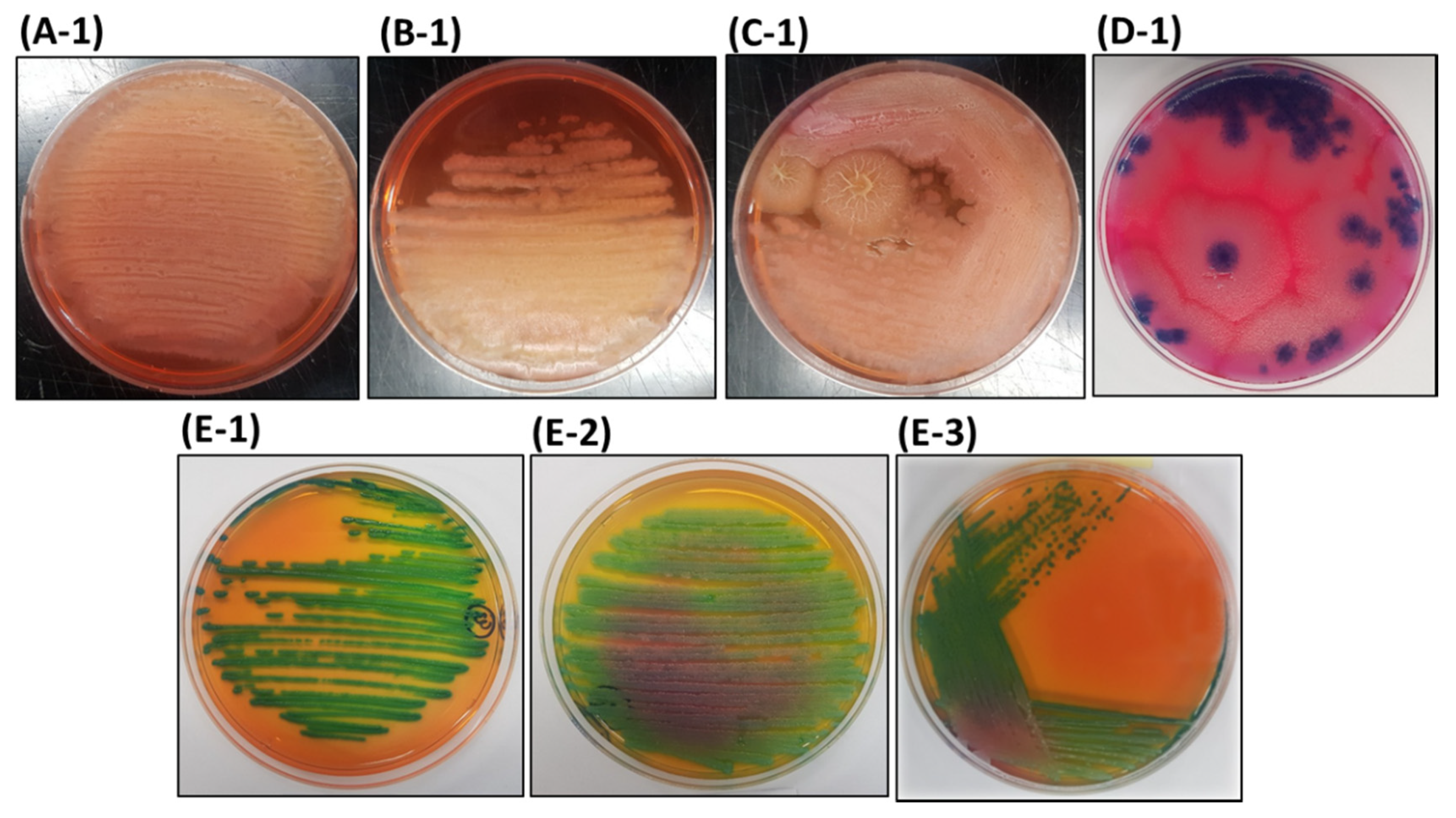
3.1. Isolation of Bacteria from AMEM
3.2. Differential Properties of the Bacterial Isolates on HiCrome™ Bacillus agar
3.3. Biochemical Characterization
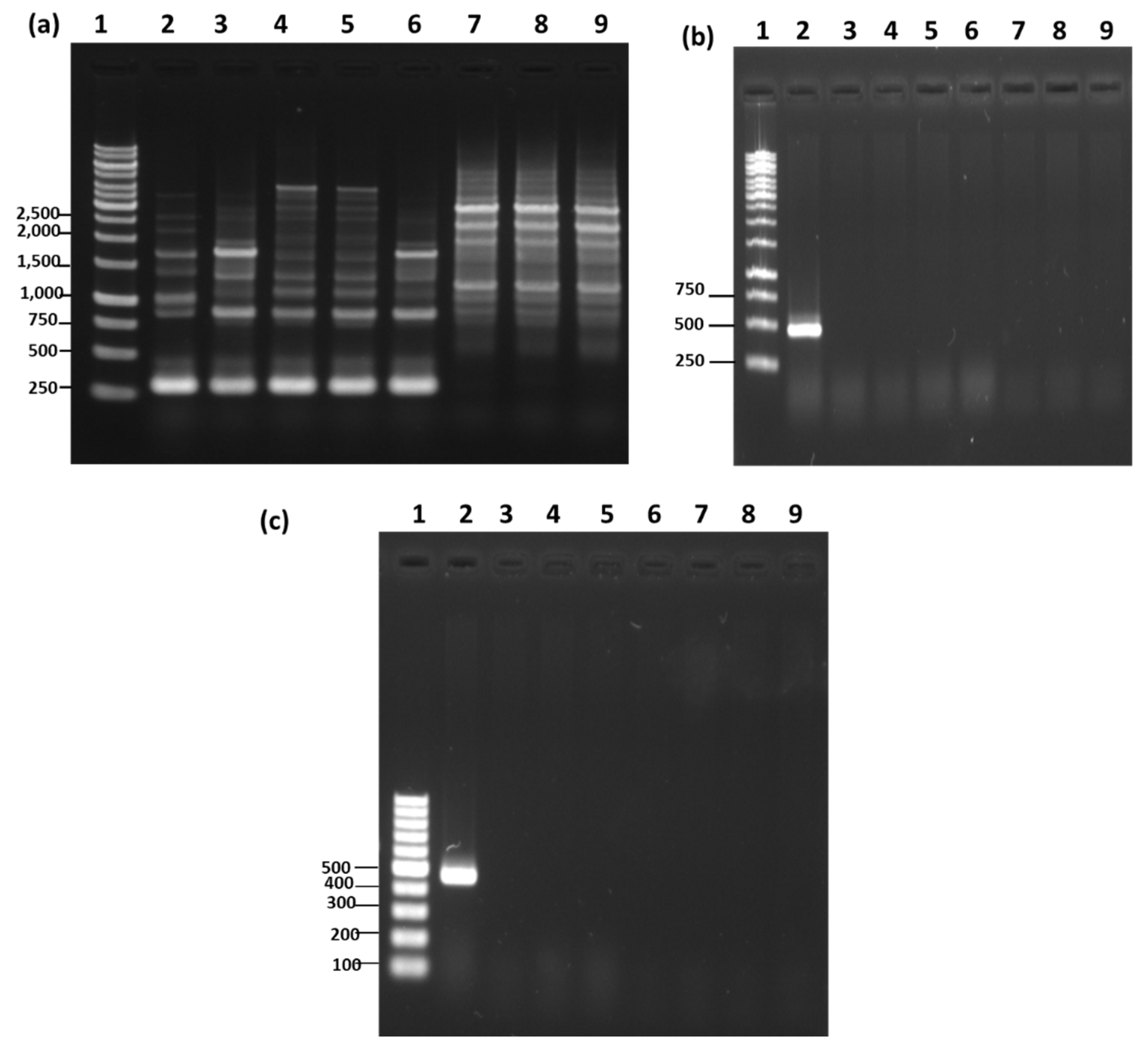
3.4. Molecular Characterization Based on 16S rRNA Gene Analysis
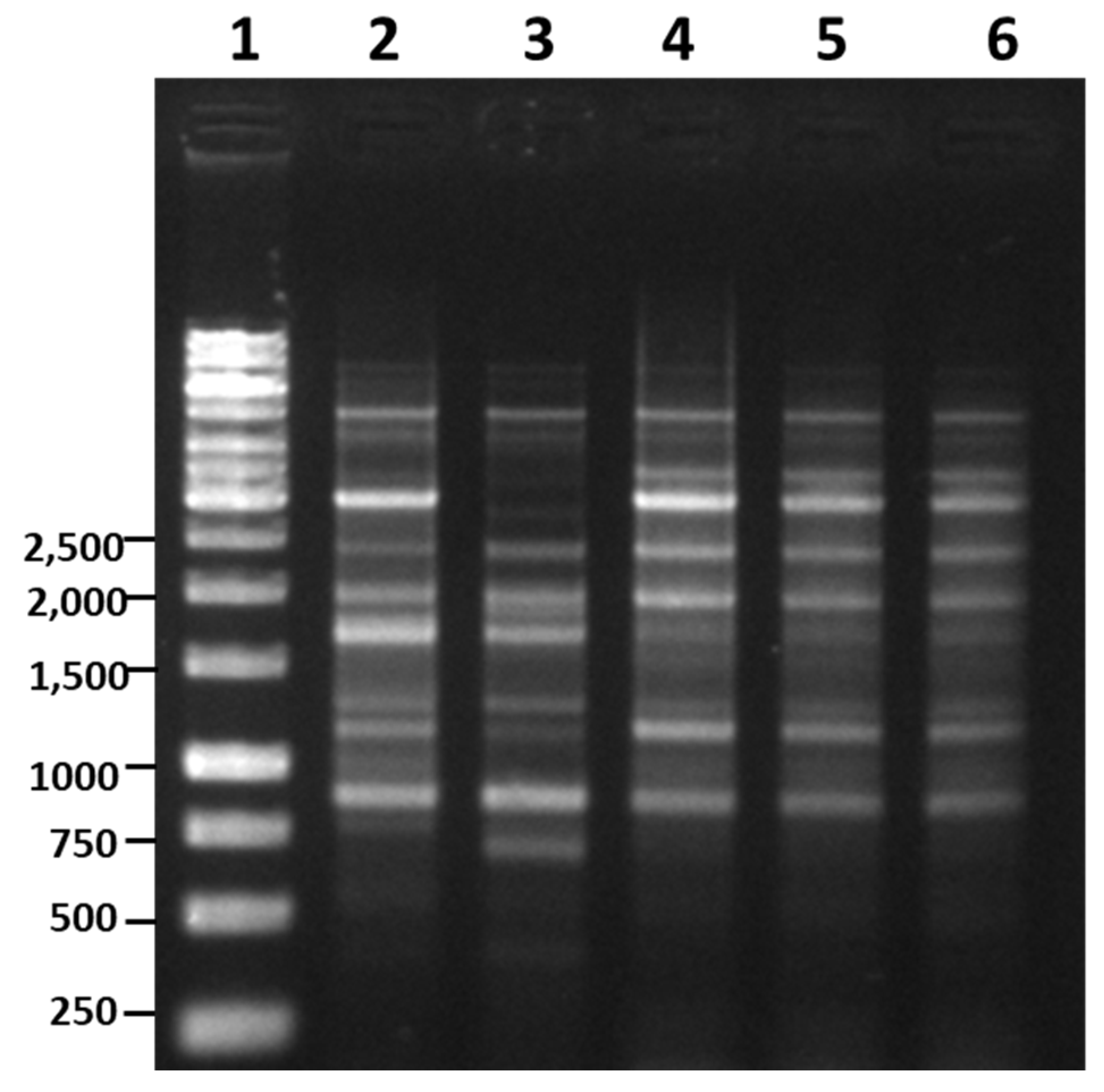
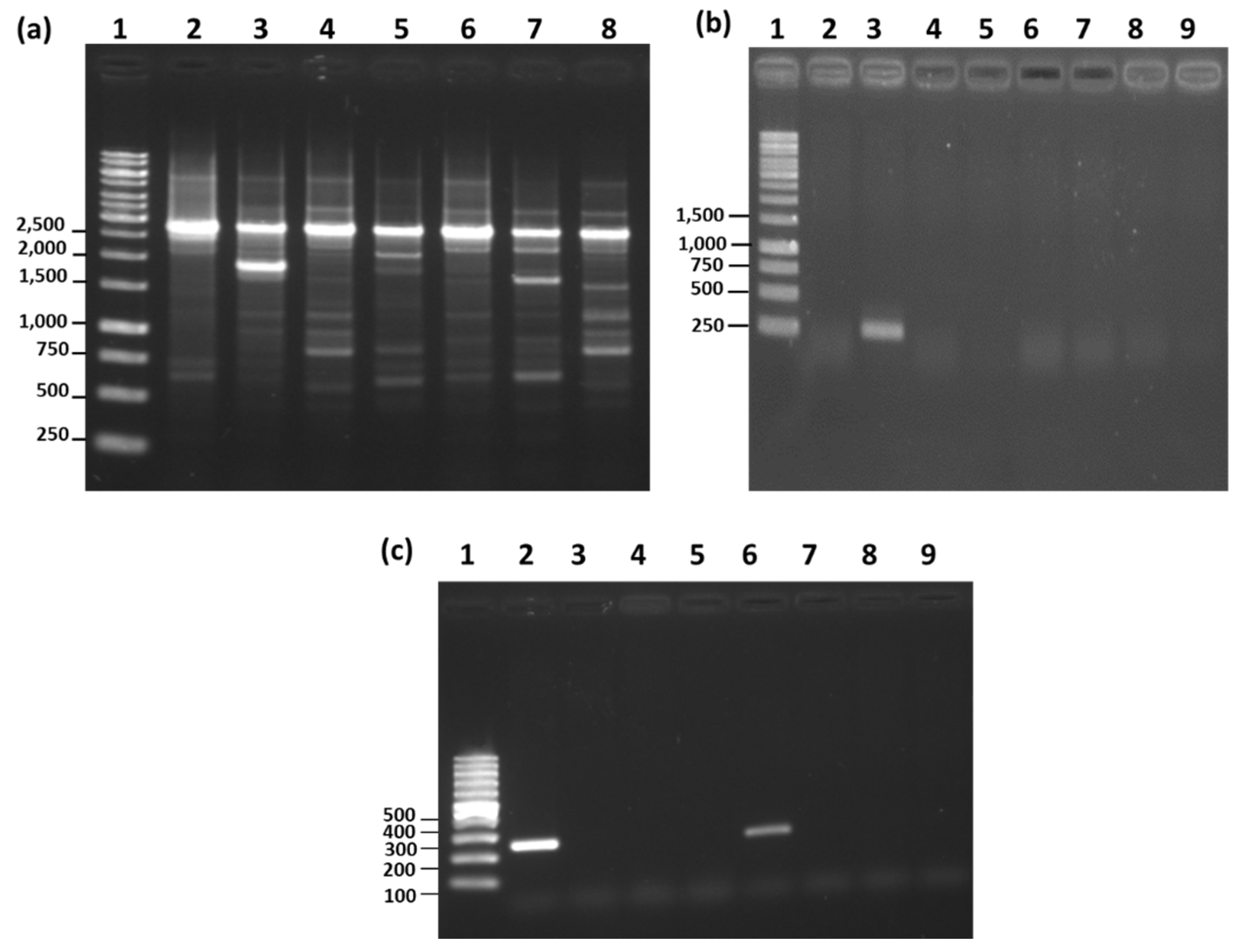
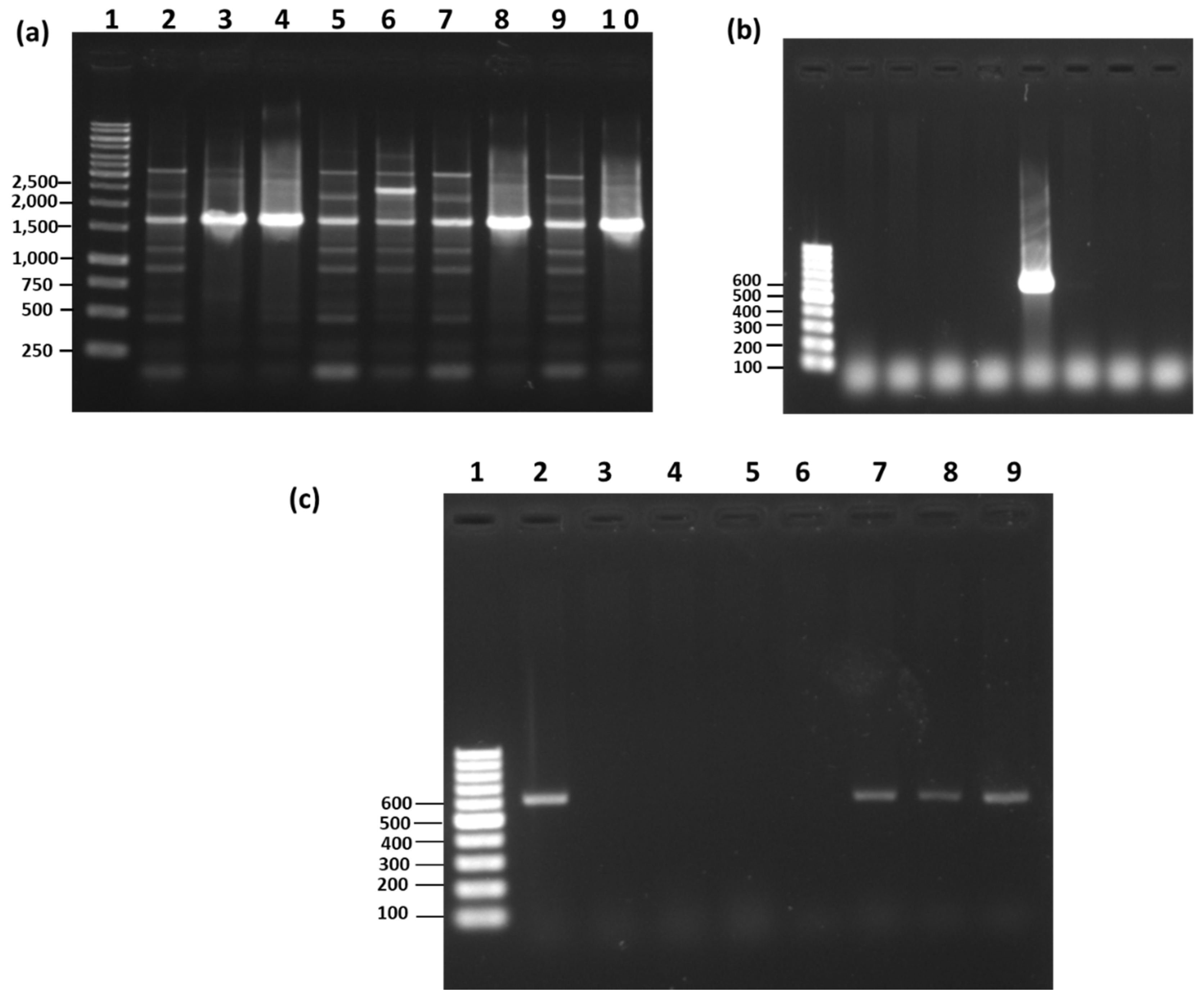
3.5. RAPD-PCR and Species-Specific PCR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, M.E. Seed and root bacterization. Annu. Rev. Phytopathol. 1974, 12, 181–197. [Google Scholar] [CrossRef]
- Schippers, B.; Scheffer, R.J.; Lugtenberg, B.J.J.; Weisbeck, P.J. Biocoating of seeds with plant growth-promoting rhizobacteria to improve plant establishment. Outlook Agric. 1995, 24, 179–185. [Google Scholar] [CrossRef]
- Pandey, A.; Tripathi, A.; Srivastava, P.; Choudhary, K.K.; Dikshit, A. Plant growth-promoting microorganisms in sustainable agriculture. In Role of Plant Growth Promoting Microorganisms in Sustainable Agriculture and Nanotechnology; Woodhead Publishing: Sawston, UK; Cambridge, UK, 2019; pp. 1–19. [Google Scholar]
- Wu, S.; Zhuang, G.; Bai, Z.; Cen, Y.; Xu, S.; Sun, H.; Zhuang, X. Mitigation of nitrous oxide emissions from acidic soils by Bacillus amyloliquefaciens, a plant growth-promoting bacterium. Glob. Chang. Biol. 2018, 24, 2352–2365. [Google Scholar] [CrossRef] [PubMed]
- Higa, T. An Earth Saving Revolution: A Means to Resolve Our World’s Problems through Effective Microorganisms (EM); Sunmark Publishing Inc.: Tokyo, Japan, 1996. [Google Scholar]
- Beall, F.; Tipping, B. Plant growth-promoting rhizobacteria in forestry. In Forest Research Marketing. Abstract 117; Ontario Forest Research Committee: Toronto, ON, Canada, 1989; p. 177. [Google Scholar]
- Holl, F.B.; Chaneway, C.P.; Turkingon, R.; Radley, R. Growth response of crested wheatgrass (Agropyron cristatum L.), white clover (Trifolium repens L.) to inoculation with Bacillus polymixa. Soil Biol. Biochem. 1988, 20, 19–24. [Google Scholar] [CrossRef]
- O’Neill, G.A.; Radley, R.A.; Chanway, C.P. Variable effects of emergence-promoting rhizobacteria on conifer seedling growth under nursery conditions. Biol. Fertil. Soils 1992, 13, 45–49. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; AbdAllah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, A.; Romero, D.; de Vicente, A. Plant protection and growth stimulation by microorganisms: Biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 2011, 22, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Ali, S.M.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Chang, M.T.; Huang, L.; Chu, W.S. Development of a novel PCR assay based on the gyrase B gene for species identification of Bacillus licheniformis. Mol. Cell. Probes 2012, 26, 215–217. [Google Scholar] [CrossRef]
- Ashe, S.; Maji, U.J.; Sen, R.; Mohanty, S.; Maiti, N.K. Specific oligonucleotide primers for detection of endoglucanase positive Bacillus subtilis by PCR. 3 Biotech 2014, 4, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Caroll, K.C.; Pfaller, M.A.; Landry, M.L.; McAdam, A.J.; Patel, R.; Richter, S.S.; Warnock, D.W. Manual of Clinical Microbiology, 12; ASM Publishing: Washington, DC, USA, 2019; p. 2668. [Google Scholar]
- Kwon, G.H.; Lee, H.A.; Park, J.Y.; Kim, J.S.; Lim, J.; Park, C.S.; Kwon, D.Y.; Kim, Y.S.; Kim, J.H. Development of a RAPD-PCR method for identification of Bacillus species isolated from Cheonggukjang. Int. J. Food Microbiol. 2009, 129, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Chelliah, R.; Park, B.J.; Kim, S.H.; Forghani, F.; Cho, M.S.; Park, D.S.; Jin, Y.G.; Oh, D.H. Differentiation of Bacillus thuringiensis from Bacillus cereus group using a unique marker based on real-time PCR. Front. Microbiol. 2019, 10, 883. [Google Scholar]
- Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-14-bacillus-cereus (accessed on 30 January 2012).
- Gotor-Vila, A.; Teixidó, N.; Usall, J.; Dashevskaya, S.; Torres, R. Development of a SCAR marker and a strain-specific genomic marker for the detection of the biocontrol agent strain CPA-8 Bacillus amyloliquefaciens (formerly B. subtilis). Ann. Appl. Biol. 2016, 169, 248–256. [Google Scholar] [CrossRef]
- Ji, Z.L.; Peng, S.; Zhu, W.; Dong, J.P.; Zhu, F. Induced resistance in nectarine fruit by Bacillus licheniformis W10 for the control of brown rot caused by Monilinia fructicola. Food Microbiol. 2020, 92, 103558. [Google Scholar] [CrossRef] [PubMed]
- Alippi, A.M.; Abrahamovich, E. HiCrome Bacillus agar for presumptive identification of Bacillus and related species isolated from honey samples. Int. J. Food Microbiol. 2019, 305, 108245. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Huang, L.; Chang, M.T.; Wu, C.P. Use of novel specific primers targeted to pheS and tuf gene for species and subspecies identification and differentiation of the Bacillus subtilis subsp. subtilis, Bacillus amyloliquefaciens and Bacillus licheniformis. Afr. J. Microbiol. Res. 2017, 11, 264–270. [Google Scholar]
- Kumar, D.; Chaudhary, K.; Boora, K.S. Characterization of native Bacillus thuringiensis strains by PCR-RAPD based fingerprinting. Indian J. Microbiol. 2010, 50, 27–32. [Google Scholar] [CrossRef] [PubMed]
| Product Code | Product Type | Product Composition | Labelled Microorganisms | pH | Aerobic Bacterial Count |
|---|---|---|---|---|---|
| AMEM-A | Liquid | -Bacillus subtilis 2% -Extracts of dried ginseng 95% | Bacillus subtilis | 5.61 ± 0.33 | 7.92 ± 0.05 Log CFU/mL |
| AMEM-B | Liquid | -Bacillus subtilis 2% -Extracts dried ginseng 60% -Extracts of cinnamon 33% | Bacillus subtilis | 6.48 ± 0.30 | 7.80 ± 0.06 Log CFU/mL |
| AMEM-C | Solid | -Bacillus subtilis culture 55.6% -Diatomite 33.4% | Bacillus subtilis | 6.67 ± 0.15 | 9.85 ± 0.08 Log CFU/g |
| AMEM-D | Solid | -Diatomite 58% -Bacillus thuringiensis 32% | Bacillus thuringiensis | 5.52 ± 0.09 | 9.30 ± 0.02 Log CFU/g |
| AMEM-E | Liquid | -Fish products 65% -Molasses 20% -Microorganisms 15% | Not specified | 3.80 ± 0.16 | 3.26 ± 0.06 Log CFU/mL |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahuguna, A.; Joe, A.-r.; Kumar, V.; Lee, J.S.; Kim, S.-Y.; Moon, J.-Y.; Cho, S.-K.; Cho, H.; Kim, M. Study on the Identification Methods for Effective Microorganisms in Commercially Available Organic Agriculture Materials. Microorganisms 2020, 8, 1568. https://doi.org/10.3390/microorganisms8101568
Bahuguna A, Joe A-r, Kumar V, Lee JS, Kim S-Y, Moon J-Y, Cho S-K, Cho H, Kim M. Study on the Identification Methods for Effective Microorganisms in Commercially Available Organic Agriculture Materials. Microorganisms. 2020; 8(10):1568. https://doi.org/10.3390/microorganisms8101568
Chicago/Turabian StyleBahuguna, Ashutosh, Ah-ryeong Joe, Vishal Kumar, Jong Suk Lee, Sung-Youn Kim, Ji-Young Moon, Soon-Kil Cho, Hyunjeong Cho, and Myunghee Kim. 2020. "Study on the Identification Methods for Effective Microorganisms in Commercially Available Organic Agriculture Materials" Microorganisms 8, no. 10: 1568. https://doi.org/10.3390/microorganisms8101568
APA StyleBahuguna, A., Joe, A.-r., Kumar, V., Lee, J. S., Kim, S.-Y., Moon, J.-Y., Cho, S.-K., Cho, H., & Kim, M. (2020). Study on the Identification Methods for Effective Microorganisms in Commercially Available Organic Agriculture Materials. Microorganisms, 8(10), 1568. https://doi.org/10.3390/microorganisms8101568





