Genome Analysis of a Marine Bacterium Halomonas sp. and Its Role in Nitrate Reduction under the Influence of Photoelectrons
Abstract
1. Introduction
2. Methods and Materials
2.1. Enrichment and Isolation of Nitrate Reducer
2.2. DNA Extraction and Sequencing
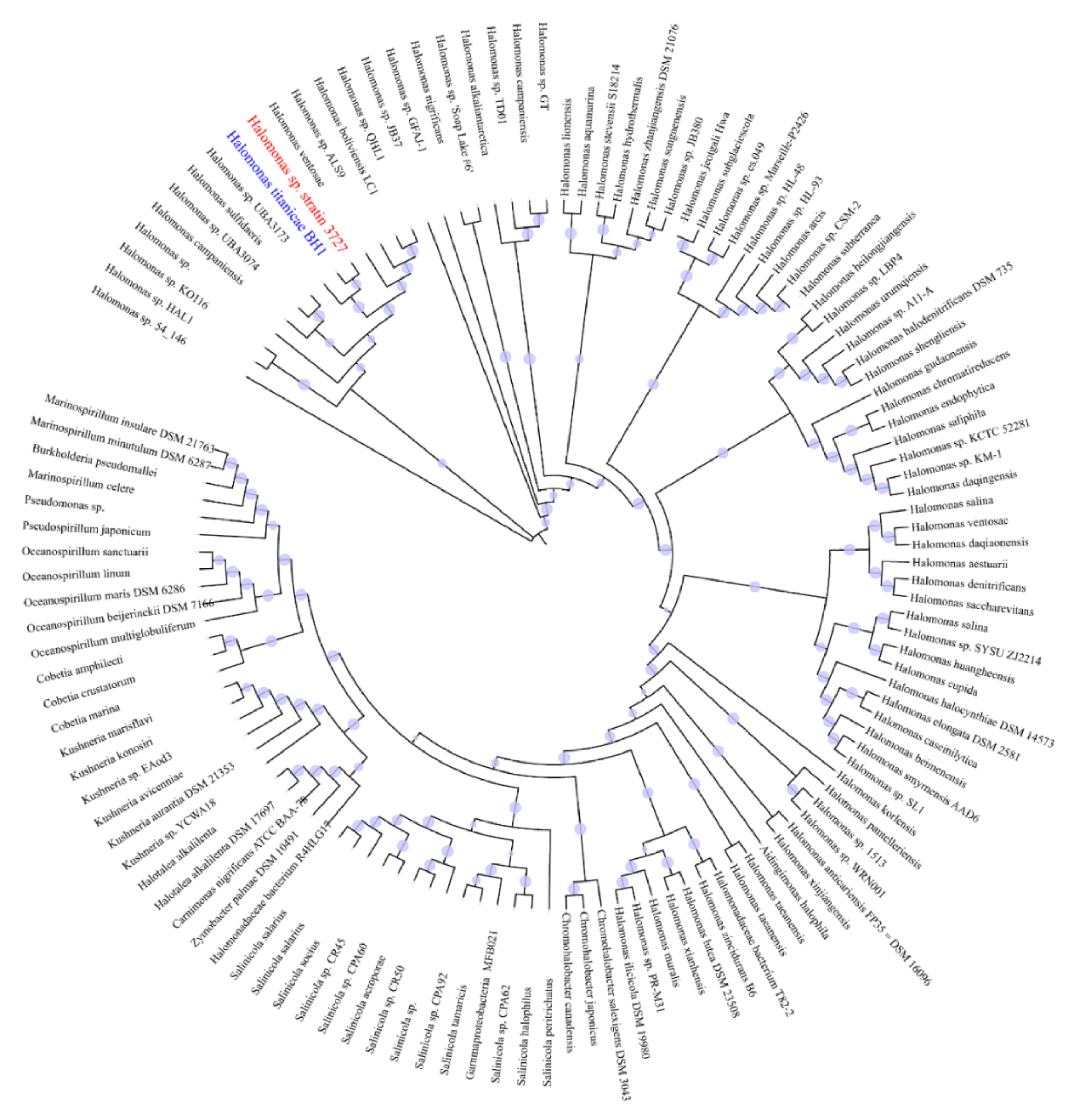
2.3. Draft Genome Sequence and Phylogenetic Analysis of Strain 3727
2.4. Nitrate and Nitrite Reduction Supplemented with Sodium Acetate
2.5. Photoelectron System Setup
2.6. Chemical Analysis Methods
3. Results and Discussion
3.1. Genome-Resolved Metagenomic and Phylogenetic Analyses
3.2. Draft Genome Features of Strain 3727
3.2.1. Carbon Metabolism
3.2.2. Nitrogen Metabolism
3.2.3. Sulfur Metabolism
3.2.4. Other Metabolisms
3.2.5. Comparison of Strain 3727, Halomonas titanicae BH1, and Halomonas UBA3171
3.3. Nitrogen Transformation Potential of Halomonas Strain 3727 under Photoelectron Impact
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Roberts, K.L.; Eate, V.M.; Eyre, B.D.; Holland, D.P.; Cook, P.L.M. Hypoxic events stimulate nitrogen recycling in a shallow salt-wedge estuary: The Yarra River estuary, Australia. Limnol. Oceanogr. 2012, 57, 1427–1442. [Google Scholar] [CrossRef]
- Woźniak, S.B.; Stramski, D. Modeling the optical properties of mineral particles suspended in seawater and their influence on ocean reflectance and chlorophyII estimation from remote sensing algorithms. Appl. Opt. 2004, 43, 3489–3503. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.J.; Tyler, C.R.; Galloway, T.S. Impacts of metal and metal oxide nanoparticles on marine organisms. Environ. Pollut. 2014, 186, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.; Raicy, M.C.; Trivedi, D.; Srinivasan, P.; Murthy, S.G.N.; Goble, R.J.; Nair, R.R. Assessment of coastal dune characteristics using georadar imaging and semdimentological analysis: Odisha and Visakhapatnam, India. J. Coast. Conserv. 2013, 17, 729–742. [Google Scholar] [CrossRef]
- Post, J.E.; Thomas, E.; Heaney, P.J. Jianshuiite in oceanic manganese nodules at the paleocene-eocene boundary. Am. Mineral. 2016, 101, 407–414. [Google Scholar] [CrossRef]
- Lu, A.; Li, Y.; Ding, H.; Xu, X.; Li, Y.; Ren, G.; Liang, J.; Liu, Y.; Hong, H.; Chen, N.; et al. Photoelectric conversion on earth’s surface via widespread Fe- and Mn-mineral coatings. Proc. Natl. Acad. Sci. USA 2019, 116, 9741–9746. [Google Scholar] [CrossRef]
- Lu, A.; Li, Y.; Jin, S.; Wang, X.; Wu, X.; Zeng, C.; Li, Y.; Ding, H.; Hao, R.; Lv, M.; et al. Growth of non-phototrophic microorganisms using solar energy through mineral photocatalysis. Nat. Commun. 2012, 3, 768. [Google Scholar] [CrossRef]
- Sakimoto, K.K.; Wong, A.B.; Yang, P.D. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 2016, 351, 74–77. [Google Scholar] [CrossRef]
- Liu, C.; Gallagher, J.J.; Sakimoto, K.K.; Nichols, E.M.; Chang, C.J.; Chang, M.C.Y.; Yang, P. Nanowire-bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals. Nano Lett. 2015, 15, 3634–3639. [Google Scholar] [CrossRef]
- Zhu, G.; Yang, Y.; Liu, J.; Liu, F.; Lu, A.; He, W. Enhanced photocurrent production by the synergy of hematite nanowire-arrayed photoanode and bioengineered Shewanella oneidensis MR-1. Biosens. Bioelectron. 2017, 94, 227–234. [Google Scholar] [CrossRef]
- Ren, G.; Yan, Y.; Sun, M.; Wang, X.; Wu, X.; Li, Y.; Lu, A.; Ding, H. Considerable bacterial community structure coupling with extracellular electron transfer at Karst area stone in Yunnan, China. Geomicrobiol. J. 2018, 35, 424–431. [Google Scholar] [CrossRef]
- Sun, M.; Ren, G.; Li, Y.; Lu, A.; Ding, H. Extracellular electron transfer between birnessite and electrochemically active bacteria community from red soil in Hainan, China. Geomicrobiol. J. 2019, 36, 169–178. [Google Scholar] [CrossRef]
- Ren, G.; Yan, Y.; Nie, Y.; Lu, A.; Wu, X.; Li, Y.; Wang, C.; Ding, H. Natural extracellular electron transfer between semiconducting minerals and electroactive bacterial communities occurred on the rock varnish. Front. Microbiol. 2019, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Koop-Jakobsen, K.; Giblin, A.E. The effect of increased nitrate loading on nitrate reduction via denitrification and DNRA in salt marsh sediments. Limnol. Oceanogr. 2010, 55, 789–802. [Google Scholar] [CrossRef]
- Tan, E.; Zou, W.; Zheng, Z.; Yan, X.; Du, M.; Hsu, T.; Tian, L.; Middelburg, J.J.; Trull, T.W.; Kao, S. Warming stimulates sediment denitrification at the expense of anaerobic ammonium oxidation. Nat. Clim. Chang. 2020, 10, 349–355. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, C.; Sheng, Y.; Dong, S.; Chen, N.; Hao, C. Effect of Fe(II) on reactivity of heterotrophic denitrifiers in the remediation of nitrate- and Fe(II)-contaminated groundwater. Ecotoxicol. Environ. Saf. 2018, 166, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sheng, Y.; Feng, C.; Chen, N.; Liu, T. Distinct functional microbial communities mediating the heterotrophic denitrification in response to the excessive Fe(II) stress in groundwater under wheat-rice stone and rock phosphate amendment. Environ. Res. 2020, 185, 109391. [Google Scholar] [CrossRef]
- Domangue, R.J.; Mortazavi, B. Nitrate reduction pathways in the presence of excess nitrogen in a shallow eutrophic estuary. Environ. Pollut. 2018, 238, 599–606. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, W.; Hou, L.; Zakem, E.J.; Wan, X.; Zhao, Z.; Liu, L.; Hunt, K.A.; Jiao, N.; Kao, S.; et al. Nitrifier adaptation to low energy flux controls inventory of reduced nitrogen in the dark ocean. Proc. Natl. Acad. Sci. USA 2020, 117, 4823–4830. [Google Scholar] [CrossRef]
- Hou, L.; Liu, M.; Carini, S.A.; Gardner, W.S. Transformation and fate of nitrate near the sediment-water interface of copano Bay. Cont. Shelf Res. 2012, 35, 86–94. [Google Scholar] [CrossRef]
- Roberson, L.A.; Kuenen, J.G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic facultatively autotrophic sulphur bacterium. J. Gen. Microbiol. 1983, 129, 2847–2855. [Google Scholar]
- Lang, X.; Li, Q.; Ji, M.; Yan, G.; Guo, S. Isolation and niche characteristics in simultaneous nitrification and denitrification application of an aerobic denitrifier, Acinetobacter sp. YS2. Bioresour. Technol. 2020, 302, 122799. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, H.; Xue, L.; Zhang, Y. Characterization of simultaneous aerobic denitrification and dephosphorization strain Cupriavidus sp. H29 and its application on cadmium-removing. Geomicrobiol. J. 2020, 37, 426–436. [Google Scholar] [CrossRef]
- Wang, X.; Yu, P.; Zeng, C.; Ding, H.; Li, Y.; Wang, C.; Lu, A. Enhanced Alcaligenes faecalis denitrification rate with electrodes as the electron donor. Appl. Environ. Microbiol. 2015, 81, 5387–5394. [Google Scholar] [CrossRef] [PubMed]
- Schut, F.; De Vries, E.; Gottschal, J.C.; Robertson, B.R.; Harder, W.; Prins, R.A.; Button, D.K. Isolation of typical marine bacteria by kilution culture: Growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 1993, 59, 2150–2160. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Wang, G.; Zhao, D.; Hao, C.; Liu, C.; Cui, L. Groundwater microbial communities along a generalized flowpath in confined aquifers in the Qaidam Basin, China. Groundwater 2018, 56, 719–731. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef]
- Tan, S.; Liu, J.; Fang, Y.; Hedlund, B.P.; Lian, Z.-H.; Huang, L.-Y.; Li, J.-T.; Huang, L.-N.; Li, W.; Jiang, H.; et al. Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 2020, 14, 1600–1613. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Posada, D., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 37, pp. 39–64. [Google Scholar]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, C.; Chen, N.; Sheng, Y.; Dong, S.; Hao, C.; Lei, K. Bioremediation of nitrate and Fe(II) combined contamination in groundwater by heterotrophic denitrifying bacteria and microbial community analysis. RSC Adv. 2016, 6, 108375–108383. [Google Scholar] [CrossRef]
- Mormile, M.R.; Edwards, T.; Frank, R.; Geurin, Z.; Haendiges, J.; Hoffmann, M.; Miller, J. Whole-genome analysis of Halomonas sp. Soap Lake#7 reveals it possesses putative Mrp antiporter operon groups 1 and 2. Genome Biol. Evol. 2019, 11, 1706–1709. [Google Scholar] [PubMed]
- Vreeland, R.H.; Litchfield, C.D.; Martin, E.L.; Elliot, E. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Evol. Microbiol. 1980, 30, 485–495. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Porro, C.; Kaur, B.; Mann, H.; Ventosa, A. Halomonas titanicae sp. nov., a halophilic bacterium isolated from the RMS Titanic. Int. J. Syst. Evol. Microbiol. 2010, 60, 2768–2774. [Google Scholar]
- Thomas, T.; Elain, A.; Bazire, A.; Bruzaud, S. Complete genome sequence of the halophilic PHS-producing bacterium Halomonas sp. SF2003: Insights into its biotechnological potential. World J. Microbiol. Biotechnol. 2019, 35, 50. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.A.; Martínez-Cánovas, J.; Quesada, E.; Béjar, V. A detailed phenotypic characterisation of the type strains of Halomonas species. Syst. Appl. Microbiol. 2002, 25, 360–375. [Google Scholar] [CrossRef]
- Voss, M.; Bange, H.W.; Dippner, J.W.; Middelburg, J.J.; Montoya, J.P.; Ward, B. The marine nitrogen cycle: Recent discoveries, uncertainties and the potential relevance of climate change. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 1621. [Google Scholar] [CrossRef]
- Narayan, K.D.; Sabat, S.C.; Das, S.K. Mechanism of electron transport during thiosulfate oxidation in an obligately mixotrophic bacterium Thiomonas bhubaneswarensis strain S10 (DSM 18181T). Appl. Microbiol. Biotechnol. 2017, 101, 1239–1252. [Google Scholar] [CrossRef]
- Kal, S.; Que, L. Dioxygen activation by nonheme iron enzymes with the 2-His-1-carboxylate facial triad that generate high-valent oxoiron oxidants. JBIC J. Biol. Inorg. Chem. 2017, 22, 339–365. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, S.; Iyer, S.R.; Babicz, J.T., Jr.; Yan, J.J.; Peters, G.H.J.; Christensen, H.E.M.; Hedman, B.; Hodgson, K.O.; Solomon, E.I. Evaluation of a concerted vs. sequential oxygen activation mechanism in α-ketoglutarate-dependent nonheme ferrous enzymes. Proc. Natl. Acad. Sci. USA 2020, 117, 5152–5159. [Google Scholar] [CrossRef] [PubMed]
- Schwibbert, K.; Marin-Sanguino, A.; Bagyan, I.; Heidrich, G.; Lentzen, G.; Seitz, H.; Rampp, M.; Schuster, S.C.; Klenk, H.-P.; Pfeiffer, F.; et al. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T: Ectoine metabolism from the Halomonas genome. Environ. Microbiol. 2011, 13, 1973–1994. [Google Scholar] [CrossRef] [PubMed]
- Williamson, A.; De Santi, C.; Altermark, B.; Karlsen, C.; Hjerde, E. Complete genome sequence of Halomonas sp. R5-57. Stand. Genom. Sci. 2016, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, Y.; Ding, H.; Ren, G.; Lu, A.; Li, Y. Photoelectron shaping marine microbial compositional and metabolic variation in the photic zone around estuary and offshore area of Yellow Sea, China. Geomicrobiol. J. 2020, 37, 716–725. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, J.J.G. How biology handle nitrite. Chem. Rev. 2014, 114, 5273–5357. [Google Scholar] [CrossRef]
- Rowley, G.; Hensen, D.; Felgate, H.; Arkenberg, A.; Appla-Ayme, C.; Prior, K.; Harrington, C.; Field, S.J.; Butt, J.N.; Baggs, E.; et al. Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar typhimurium. Biochem. J. 2012, 441, 755–762. [Google Scholar] [CrossRef]
- Gilberthorpe, N.J.; Poole, R.K. Nitric oxide homeostasis in salmonella typhimurium. J. Biol. Chem. 2008, 283, 11146–11154. [Google Scholar] [CrossRef]



| Potential | NO3−-N | NO2−-N | NH4+-N | N Loss in Liquid Phase | Faradaic Efficiency |
|---|---|---|---|---|---|
| −0.15V | 21.20% | 51.36% | 6.77% | 20.67% | 25.28% |
| −0.20V | 0.00% | 68.40% | 8.52% | 23.08% | 4.28% |
| −0.30V | 40.74% | 30.03% | 3.77% | 25.19% | 3.82% |
| Open Circuit | 63.94% | 26.77% | 11.17% | 0.00% | / |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ding, H.; Sun, Y.; Li, Y.; Lu, A. Genome Analysis of a Marine Bacterium Halomonas sp. and Its Role in Nitrate Reduction under the Influence of Photoelectrons. Microorganisms 2020, 8, 1529. https://doi.org/10.3390/microorganisms8101529
Liu Y, Ding H, Sun Y, Li Y, Lu A. Genome Analysis of a Marine Bacterium Halomonas sp. and Its Role in Nitrate Reduction under the Influence of Photoelectrons. Microorganisms. 2020; 8(10):1529. https://doi.org/10.3390/microorganisms8101529
Chicago/Turabian StyleLiu, Ying, Hongrui Ding, Yuan Sun, Yan Li, and Anhuai Lu. 2020. "Genome Analysis of a Marine Bacterium Halomonas sp. and Its Role in Nitrate Reduction under the Influence of Photoelectrons" Microorganisms 8, no. 10: 1529. https://doi.org/10.3390/microorganisms8101529
APA StyleLiu, Y., Ding, H., Sun, Y., Li, Y., & Lu, A. (2020). Genome Analysis of a Marine Bacterium Halomonas sp. and Its Role in Nitrate Reduction under the Influence of Photoelectrons. Microorganisms, 8(10), 1529. https://doi.org/10.3390/microorganisms8101529





