Long-Term Helicobacter pylori Infection Switches Gastric Epithelium Reprogramming towards Cancer Stem Cell-Related Differentiation Program in Hp-Activated Gastric Fibroblast-TGFβ Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Hp Strain
2.2. Technique of Rat Gastric Fibroblasts Isolation and Their Activation towards Fibroblasts Possessing CAFs Characteristic
2.3. Isolation of Hp-AGF and Normal Gastric Fibroblast (GF)-Conditioned Media
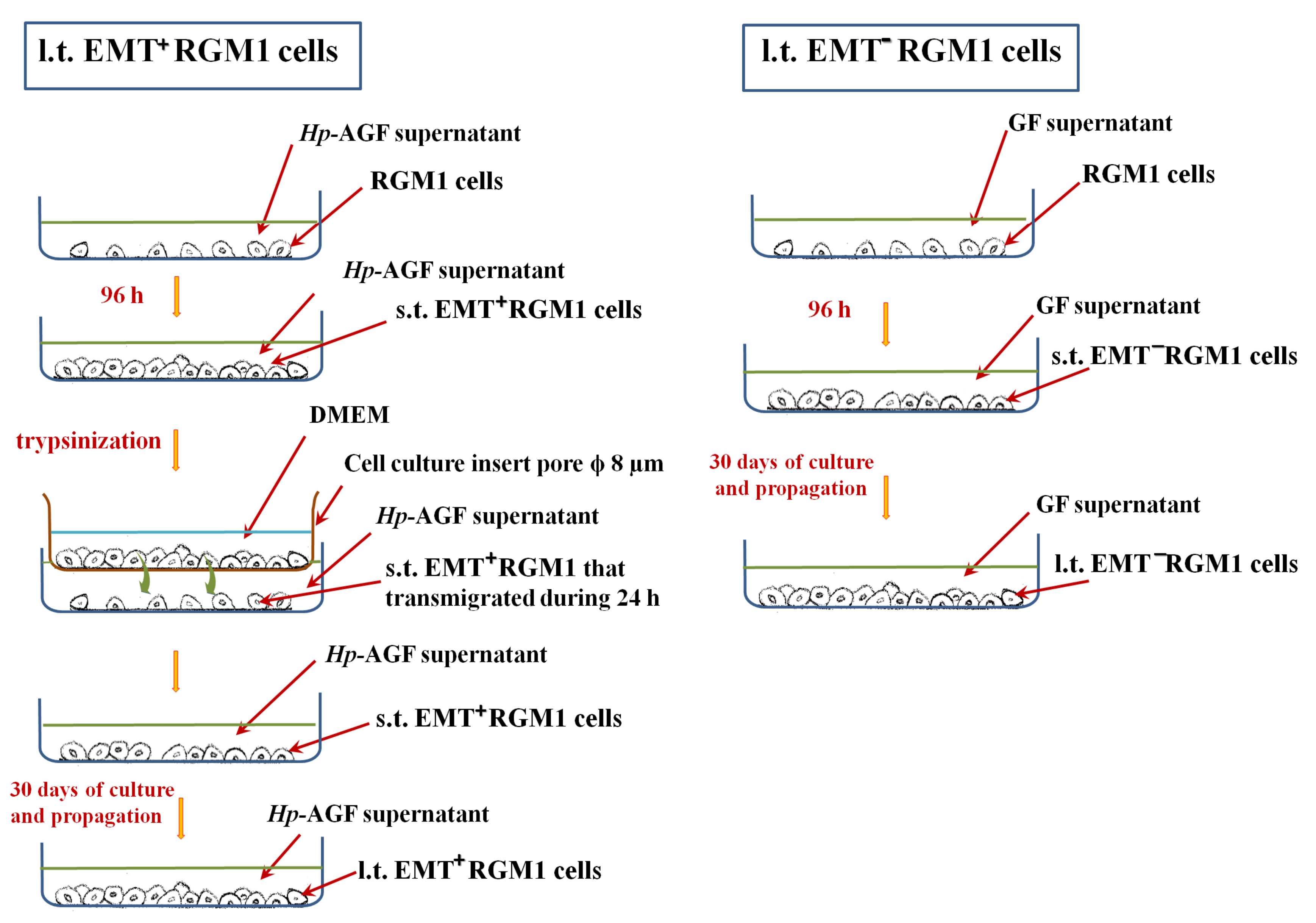
2.4. Long-Term Influence of Supernatants Collected from Hp-AGFs and GFs on Epithelial RGM1 Cells
2.5. RT-PCR Technique
2.6. Influence of TGFβR1 Kinase Activity Receptor Inhibition on Phenotype of Long-Term RGM1 Cells
2.7. Cell Proliferation
2.8. Western Blot
2.9. Determination of the Cells Ability to Release TGFβ1
2.10. Image Acquisition and Immunofluorescence
2.11. Statistical Analyses
3. Results
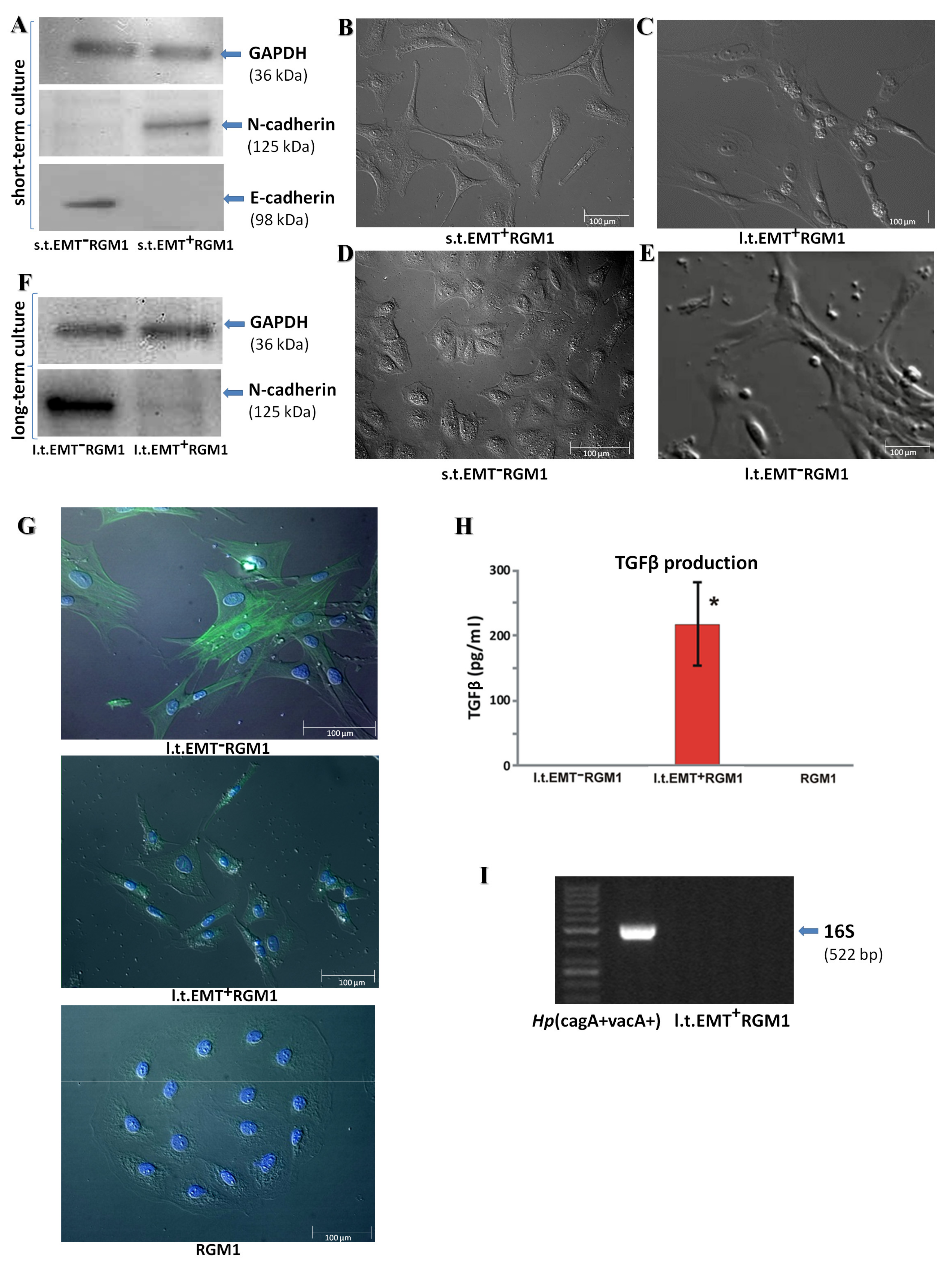
3.1. Long-Term Influence of Hp-AGF Secretome Induces Phenotypical Changes in Pro-Invasive s.t.EMT+RGM1 Cells
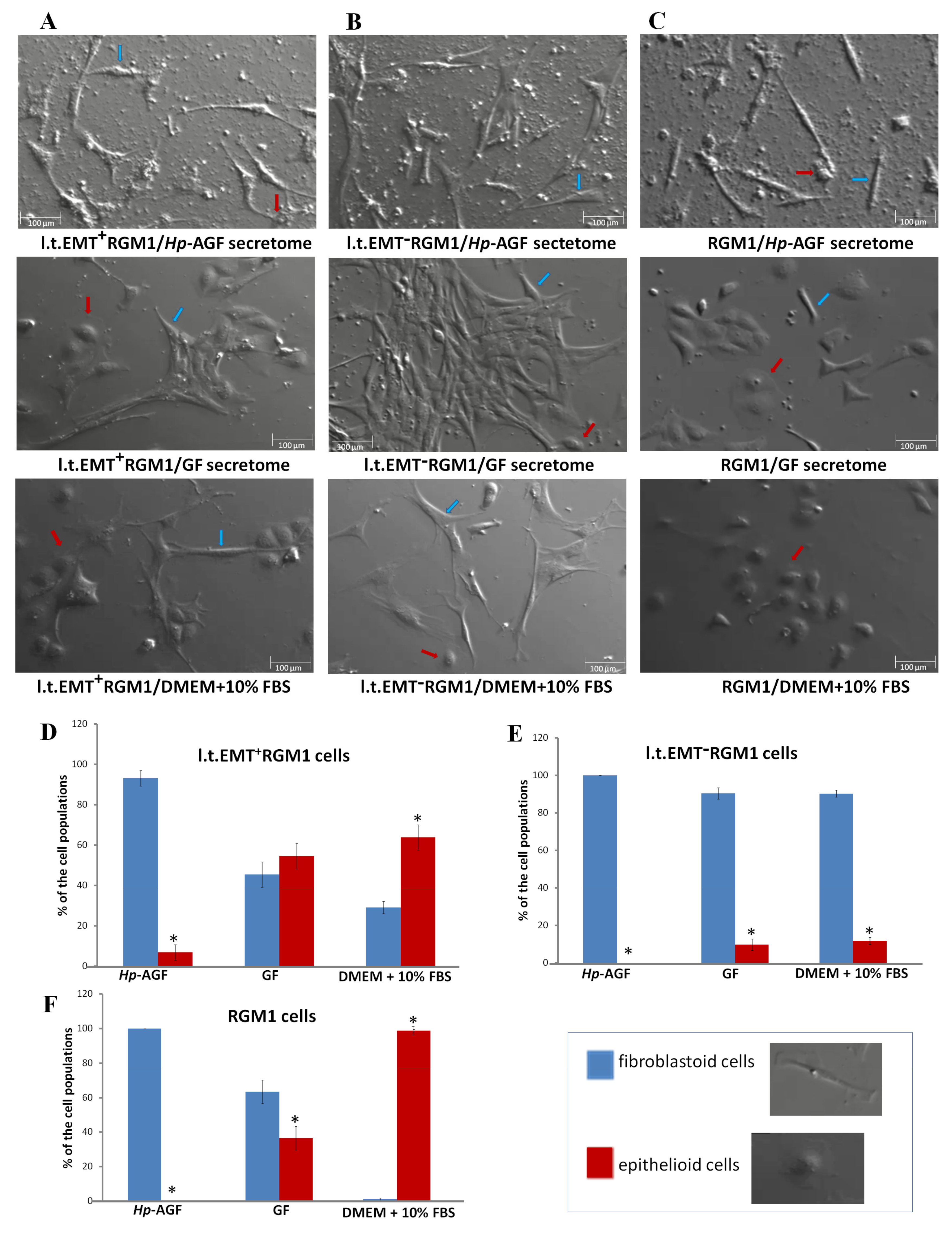
3.2. The Phenotypic Plasticity of l.t.EMT+RGM1 Cells
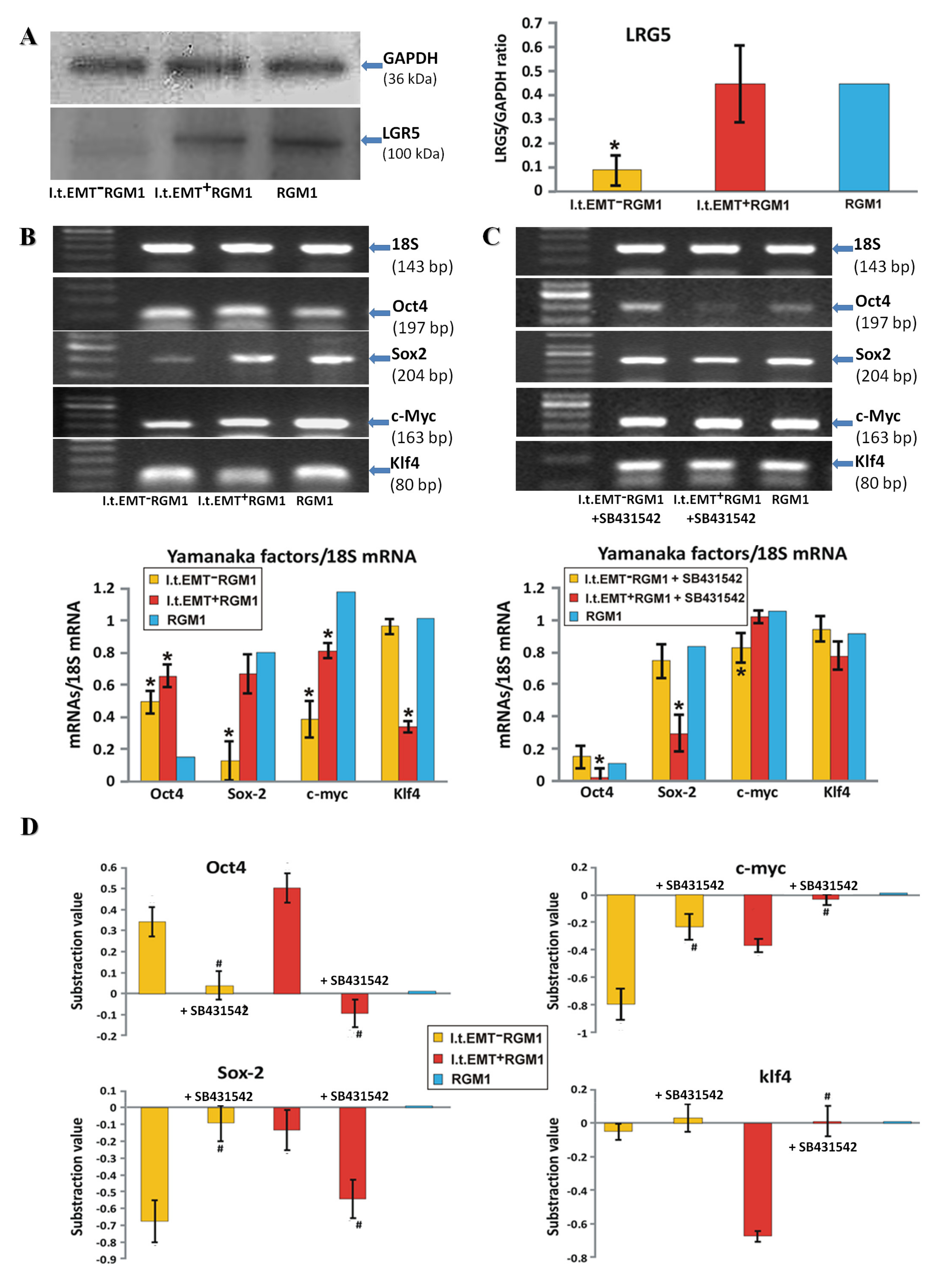
3.3. TGFβR1 Activation Participates in Both Pro-Pluripotent and Pro-Fibrotic RGM1 Reprogramming
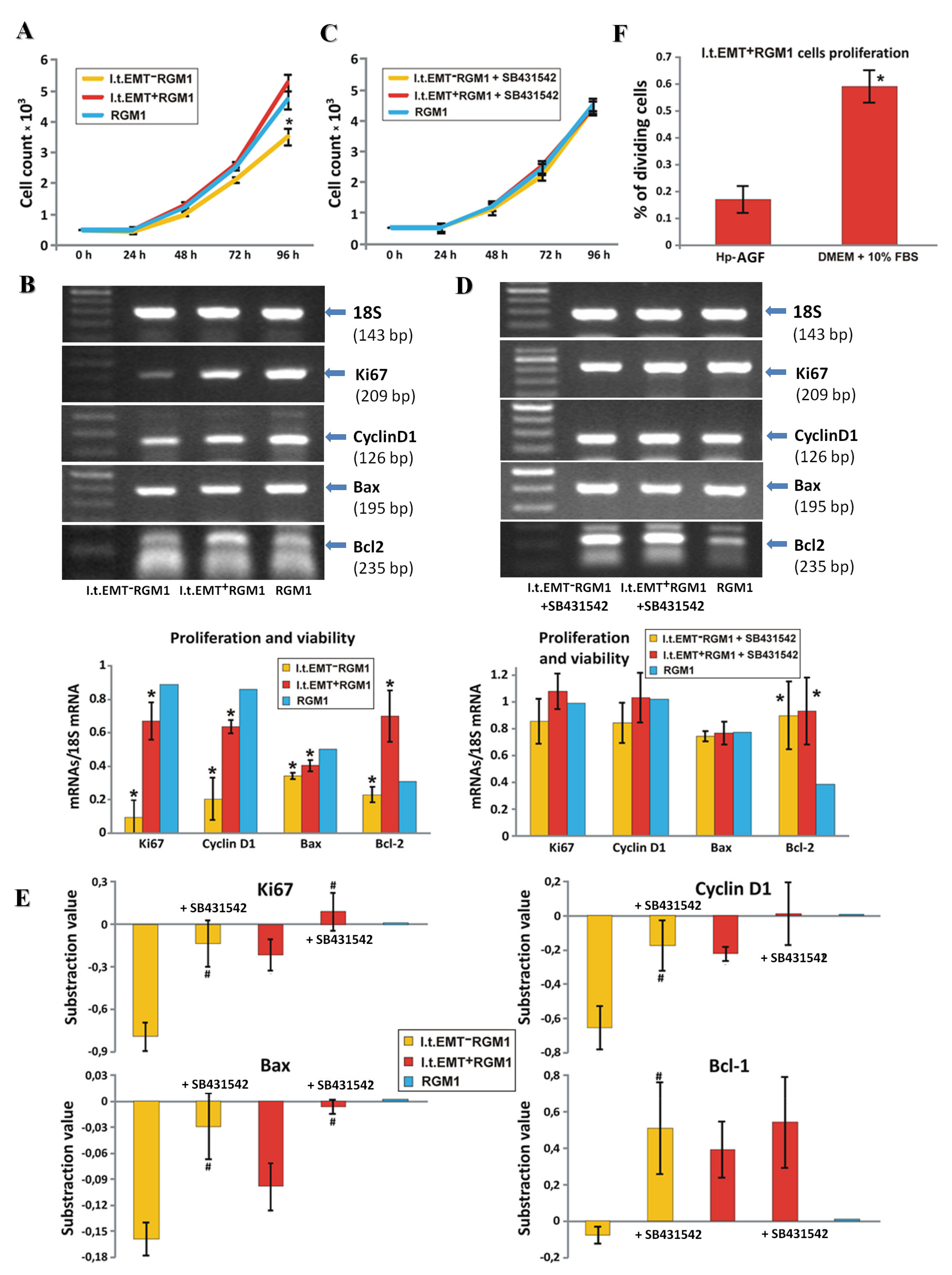
3.4. Hp-AGF Secretome Induces Potentially Pro-Proliferative Phenotype of l.t.EMT+RGM1 Cells
3.5. Hp-AGF Secretome Prompts TGFβ-Dependent Proliferation of l.t.EMT+RGM1 Cells
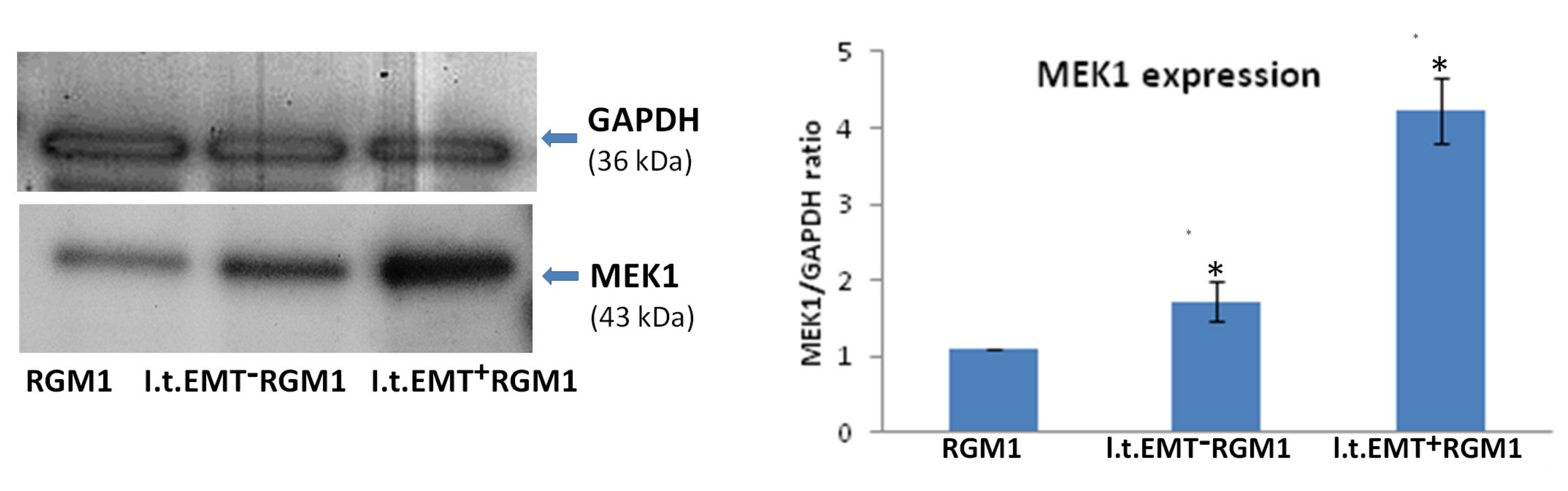
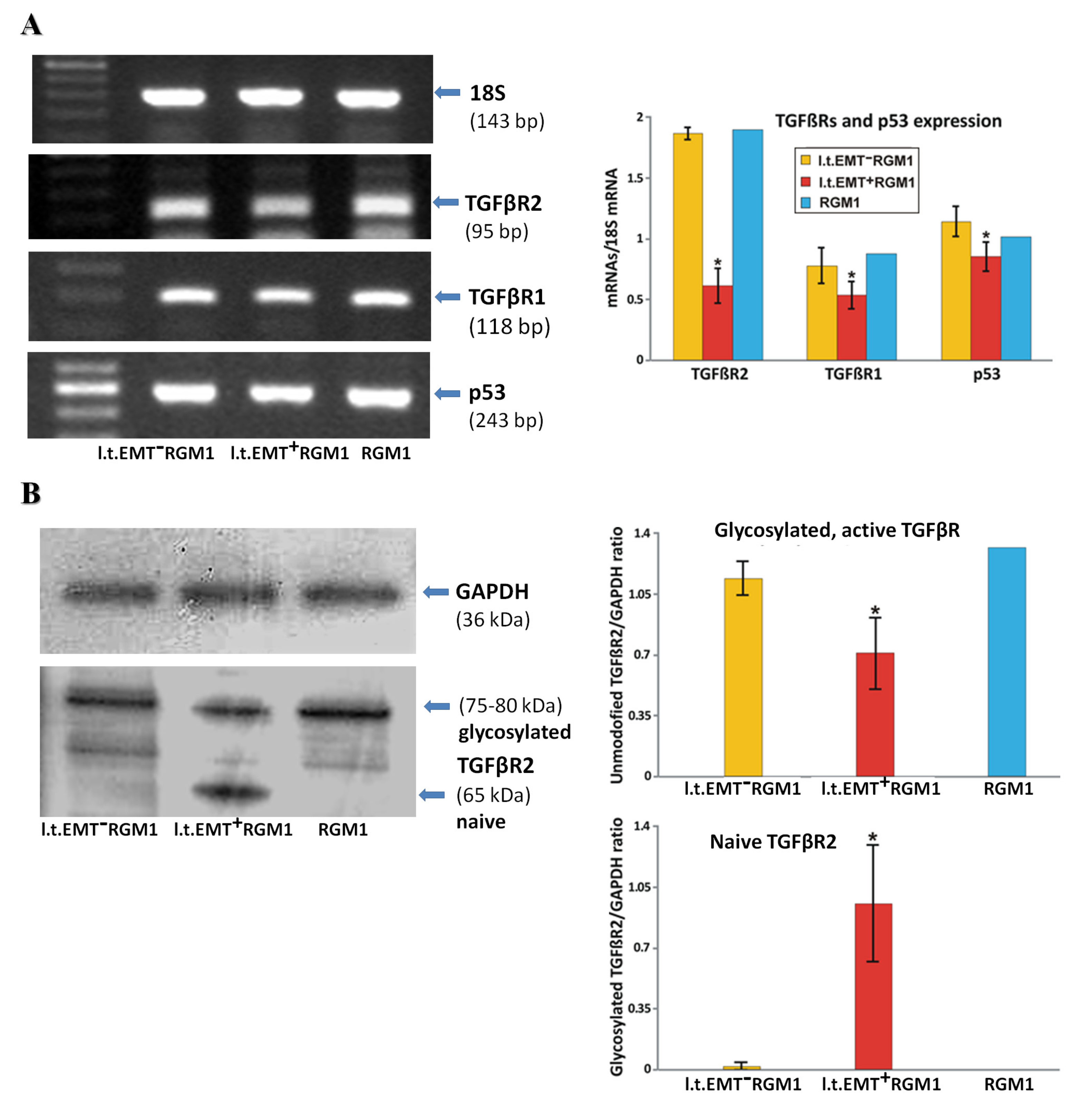
3.6. Interrelations between TGFβR1 and TGFβR2 Activity Underlie Differential Microevolution Pattern of l.t.EMT+RGM1 and l.t.EMT−RGM1 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dicken, B.J.; Bigam, D.L.; Cass, C.; Mackey, J.R.; Joy, A.A.; Hamilton, S.M. Gastric adenocarcinoma: Review and considerations for future directions. Ann. Surg. 2005, 241, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kong, Y.; Weng, S.; Dong, C.; Zhu, L.; Yang, Z.; Zhong, J.; Yuan, Y. Outcomes of surgery for gastric cancer with distant metastases: A retrospective study from the SEER database. Oncotarget 2017, 8, 4342–4351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apicella, M.; Corso, S.; Giordano, S. Targeted therapies for gastric cancer: Failures and hopes from clinical trias. Oncotarget 2017, 8, 57654–57669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavarría-Velázquez, C.O.; Torres-Martínez, A.C.; Montaño, L.F.; Rendón-Huerta, E.P. TLR2 Activation induced by H. pylori LPS promotes the differential expression of claudin-4-6-7 and -9 via either STAT3 and ERK1/2 in AGS cells. Immunobiology 2018, 223, 38–48. [Google Scholar] [CrossRef] [PubMed]
- López-Novoa, J.M.; Nieto, M.A. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 2009, 1, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Posselt, G.; Backert, S.; Wessler, S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun. Signal. 2013, 11, 77. [Google Scholar] [CrossRef] [Green Version]
- Schneider, S.; Carra, G.; Sahin, U.; Hoy, B.; Rieder, G.; Wessler, S. Complex cellular responses of Helicobacter pylori-colonized gastric adenocarcinoma cells. Infect. Immun. 2011, 79, 2362–2371. [Google Scholar] [CrossRef] [Green Version]
- Wessler, S.; Backert, S. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 2008, 16, 397–405. [Google Scholar] [CrossRef]
- Wang, F.; Meng, W.; Wang, B.; Qiao, L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014, 345, 196–202. [Google Scholar] [CrossRef]
- Ishimoto, T.; Sawayama, H.; Sugihara, H.; Baba, H. Interaction between gastric cancer stem cells and the tumor microenvironment. J. Gastroenterol. 2014, 49, 1111–1120. [Google Scholar] [CrossRef]
- Chung, H.W.; Lim, J.B. Role of the tumor microenvironment in the pathogenesis of gastric carcinoma. World J. Gastroenterol. 2014, 20, 1667–1680. [Google Scholar] [CrossRef] [PubMed]
- Necchi, V.; Candusso, M.; Tava, F.; Luinetti, O.; Ventura, U.; Fiocca, R.; Ricci, V.; Solcia, E. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology 2007, 132, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.L.; Hsu, W.H.; Kao, M.C.; Chou, C.C.; Lin, C.C.; Liu, C.J.; Weng, B.C.; Kuo, F.C.; Kuo, C.H.; Lin, M.H.; et al. Stromal C-type lectin receptor COLEC12 integrates H. pylori, PGE2-EP2/4 axis and innate immunity in gastric diseases. Sci. Rep. 2018, 8, 3821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, M.; Blobe, G.C. Role of transforming growth factor-beta in hematologic malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef] [Green Version]
- Spender, L.C.; O’Brien, D.I.; Simpson, D.; Dutt, D.; Gregory, C.D.; Allday, M.J.; Clark, L.J.; Inman, G.J. TGF-β induces apoptosis in human B cells via transcriptional regulation of BIK and BCL-XL. Cell Death Differ. 2009, 16, 593–602. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [Green Version]
- Massagué, J. TGFβ in cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [Green Version]
- Ham, I.H.; Lee, D.; Hur, H. Role of cancer-associated fibroblast in gastric cancer progression and resistance to treatments. J. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Paraiso, K.H.; Smalley, K.S. Fibroblast-Mediated drug resistance in cancer. Biochem. Pharmacol. 2013, 85, 1033–1041. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Sakai, R. Direct interaction between carcinoma cells and cancer associated fibroblasts for the regulation of cancer invasion. Cancers 2015, 7, 2054–2062. [Google Scholar] [CrossRef] [PubMed]
- Krzysiek-Maczka, G.; Wrobel, T.; Targosz, A.; Szczyrk, U.; Strzalka, M.; Ptak-Belowska, A.; Czyz, J.; Brzozowski, T. Helicobacter pylori-activated gastric fibroblasts induce epithelial-mesenchymal transition of gastric epithelial cells in vitro in a TGF-β-dependent manner. Helicobacter 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Kuzet, S.E.; Gaggioli, C. Fibroblast activation in cancer: When seed fertilizes soil. Cell Tissue Res. 2016, 365, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, K.; Murata, M.; Yamaguchi, T.; Matsuzaki, K.; Okazaki, K. Reversible human TGF-β signal shifting between tumor suppression and fibro-carcinogenesis: Implications of Smad phospho-isoforms for hepatic epithelial-mesenchymal transitions. J. Clin. Med. 2016, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.; Moon, A. Inflammatory fibroblasts in cancer. Arch. Pharm. Res. 2016, 39, 1021–1031. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, G.; Wang, B.; Wu, J.; Cao, Y.; Zhu, D.; Xu, Y.; Wang, X.; Han, H.; Li, X.; et al. Cancer-Associated fibroblasts promote irradiated cancer cell recovery through autophagy. EBioMedicine 2017, 17, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.T.; Sun, W.; Zhang, J.T.; Fan, Y.Z. Cancer-Associated fibroblast regulation of tumor neo-angiogenesis as a therapeutic target in cancer. Oncol. Lett. 2019, 17, 3055–3065. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.X.; Guan, X.Y.; Fu, L. Therapeutic targeting of the crosstalk between cancer-associated fibroblasts and cancer stem cells. Am. J. Cancer Res. 2019, 9, 1889–1904. [Google Scholar]
- Müller, M.; Hermann, P.C.; Liebau, S.; Weidgang, C.; Seufferlein, T.; Kleger, A.; Perkhofer, L. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 2016, 16, 349–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aponte, P.M.; Caicedo, A. Stemness in cancer: Stem cells, cancer stem cells, and their microenvironment. Stem Cells Int. 2017, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, M.C.; Hollingsworth, R.E.; Hurt, E.M. Cancer stem cell plasticity and tumor hierarchy. World J. Stem Cells 2015, 7, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Jiang, J.; Liang, X.H.; Tang, T.L. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther. 2015, 8, 2973–2980. [Google Scholar] [PubMed] [Green Version]
- Li, C.J.; Chu, P.Y.; Yiang, G.T.; Wu, M.Y. The molecular mechanism of epithelial-mesenchymal transition for breast carcinogenesis. Biomolecules 2019, 9, 476. [Google Scholar] [CrossRef] [Green Version]
- Krzysiek-Maczka, G.; Targosz, A.; Szczyrk, U.; Strzalka, M.; Brzozowski, T.; Ptak-Belowska, A. Involvement of epithelial-mesenchymal transition-inducing transcription factors in the mechanism of Helicobacter pylori-induced fibroblasts activation. J. Physiol. Pharmacol. 2019, 70. [Google Scholar] [CrossRef]
- Krzysiek-Maczka, G.; Targosz, A.; Ptak-Belowska, A.; Korbut, E.; Szczyrk, U.; Strzalka, M.; Brzozowski, T. Molecular alterations in fibroblasts exposed to Helicobacter pylori: A missing link in bacterial inflammation progressing into gastric carcinogenesis? J. Physiol. Pharmacol. 2013, 64, 77–87. [Google Scholar] [PubMed]
- Krzysiek-Maczka, G.; Targosz, A.; Szczyrk, U.; Strzałka, M.; Sliwowski, Z.; Brzozowski, T.; Czyz, J.; Ptak-Belowska, A. Role of Helicobacter pylori infection in cancer-associated fibroblast- induced epithelial-mesenchymal transition in vitro. Helicobacter 2018, 23. [Google Scholar] [CrossRef]
- Kobayashi, I.; Kawano, S.; Tsuji, S.; Matsui, H.; Nakama, A.; Sawaoka, H.; Masuda, E.; Takei, Y.; Kouichi Nagano, K.; Fusamoto, H.; et al. RGM1, a cell line derived from normal gastric mucosa of rat. In Vitro Cell Dev. Biol. Anim. 1996, 32, 259–261. [Google Scholar] [CrossRef]
- Lu, H.J.; Yan, J.; Jin, P.Y.; Zheng, G.H.; Qin, S.M.; Wu, D.M.; Lu, J.; Zheng, Y.L. MicroRNA-152 inhibits tumor cell growth while inducing apoptosis via the transcriptional repression of cathepsin L in gastrointestinal stromal tumor. Cancer Biomark. 2018, 21, 711–722. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-Step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.Y.; Ooshima, A.; Yang, K.M.; Kang, J.M.; Kim, Y.W.; Kim, S.J. Glucosamine hydrochloride exerts a protective effect against unilateral ureteral obstruction-induced renal fibrosis by attenuating TGF-β signaling. J. Mol. Med. 2013, 91, 1273–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marte, B. Tumour heterogeneity. Nature 2013, 501, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brungs, D.; Aghmesheh, M.; Vine, K.L.; Becker, T.M.; Carolan, M.G.; Ranson, M. Gastric cancer stem cells: Evidence, potential markers, and clinical implications. J. Gastroenterol. 2016, 51, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Bekaii-Saab, T.; El-Rayes, B. Identifying and targeting cancer stem cells in the treatment of gastric cancer. Cancer 2017, 123, 1303–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May, C.D.; Sphyris, N.; Evans, K.W.; Werden, S.J.; Guo, W.; Mani, S.A. Epithelial-Mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011, 13, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maugeri-Saccà, M.; Vigneri, P.; De Maria, R. Cancer stem cells and chemosensitivity. Clin. Cancer Res. 2011, 17, 4942–4947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochedlinger, K.; Yamada, Y.; Beard, C.; Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005, 121, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Al-Marzoqee, F.Y.; Khoder, G.; Al-Awadhi, H.; John, R.; Beg, A.; Vincze, A.; Branicki, F.; Karam, S.M. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int. J. Oncol. 2012, 41, 1733–1743. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, W.R.; Qian, H.; Zhu, W.; Bu, X.B.; Wang, S.; Yan, Y.M.; Mao, F.; Gu, H.B.; Cao, H.L.; et al. Oct4, a novel marker for human gastric cancer. J. Surg. Oncol. 2009, 99, 414–419. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S.; Zhang, L. Upregulated miR-132 in Lgr5(+) gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Hoffmann, W. Current status on stem cells and cancers of the gastric epithelium. Int. J. Mol. Sci. 2015, 16, 19153–19169. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; Todaro, M.; De Sousa Melo, F.; Sprick, M.R.; Kemper, K.; Perez Alea, M.; Richel, D.J.; Stassi, G.; Medema, J.P. Single-Cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA 2008, 105, 13427–13432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, N.M.; Maddipati, R.; Norgard, R.J.; Balli, D.; Li, J.; Yuan, S.; Yamazoe, T.; Black, T.; Sahmoud, A.; Furth, E.E.; et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 2018, 45, 681–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, N.M.; Kang, Y. Context-Dependent EMT programs in cancer metastasis. J. Exp. Med. 2019, 216, 1016–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, Y.; Mou, Z.; Chen, J.; He, Y.; Dong, H.; Fu, X.; Wu, Y. B7H1 expression and epithelial- to-mesenchymal transition phenotypes on colorectal cancer stem-like cells. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.W.; Su, Y.J.; Hsiao, M.; Wei, K.C.; Lin, W.H.; Liang, C.L.; Chen, S.C.; Lee, J.L. Diverse targets of β-catenin during the epithelial-mesenchymal transition define cancer stem cells and predict disease relapse. Cancer Res. 2015, 75, 3398–3410. [Google Scholar] [CrossRef] [Green Version]
- Garg, M. Urothelial cancer stem cells and epithelial plasticity: Current concepts and therapeutic implications in bladder cancer. Cancer Metastasis Rev. 2015, 34, 691–701. [Google Scholar] [CrossRef]
- Unternaehrer, J.J.; Zhao, R.; Kim, K.; Cesana, M.; Powers, J.T.; Ratanasirintrawoot, S.; Onder, T.; Shibue, T.; Weinberg, R.A.; Daley, G.Q. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep. 2014, 3, 691–698. [Google Scholar] [CrossRef]
- Comes, S.; Gagliardi, M.; Laprano, N.; Fico, A.; Cimmino, A.; Palamidessi, A.; De Cesare, D.; De Falco, S.; Angelini, A.; Scita, G.; et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Rep. 2013, 1, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonardo, E.; Hermann, P.C.; Mueller, M.T.; Huber, S.; Balic, A.; Miranda-Lorenzo, I.; Zagorac, S.; Alcala, S.; Rodriguez-Arabaolaza, I.; Carlos Ramirez, J. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 2011, 9, 433–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sainz, B., Jr.; Alcala, S.; Garcia, S.; Sanchez-Ripoll, Y.; Azevedo, M.M.; Cioffi, M.; Tatari, M.; Miranda-Lorenzo, I.; Hidalgo, M.; Gomez-Lopez, G.; et al. Microenvironmental hCAP-18/LL-37 promotes pancreatic ductal adenocarcinoma by activating its cancer stem cell compartment. Gut 2015, 64, 1921–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermann, P.C.; Sancho, P.; Canamero, M.; Martinelli, P.; Madriles, F.; Michl, P.; Gress, T.; De Pascual, R.; Gandia, L.; Guerra, C.; et al. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterology 2014, 147. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.C.; Trabulo, S.M.; Sainz, B., Jr.; Balic, A.; Garcia, E.; Hahn, S.A.; Vandana, M.; Sahoo, S.K.; Tunici, P.; Bakker, A.; et al. Multimodal treatment eliminates cancer stem cells and leads to long-term survival in primary human pancreatic cancer tissue xenografts. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Mueller, M.T.; Hermann, P.C.; Witthauer, J.; Rubio-Viqueira, B.; Leicht, S.F.; Huber, S.; Ellwart, J.W.; Mustafa, M.; Bartenstein, P.; D’Haese, J.G.; et al. Combined targeted treatment to eliminate tumorigenic cancer stem cells in human pancreatic cancer. Gastroenterology 2009, 137, 1102–1113. [Google Scholar] [CrossRef]
- Radisky, D.C.; LaBarge, M.A. Epithelial-Mesenchymal transition and the stem cell phenotype. Cell Stem Cell 2008, 2, 511–512. [Google Scholar] [CrossRef] [Green Version]
- Scheel, C.; Weinberg, R.A. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int. J. Cancer 2011, 129, 2310–2314. [Google Scholar] [CrossRef] [Green Version]
- Giannoni, E.; Bianchini, F.; Masieri, L.; Serni, S.; Torre, E.; Calorini, L.; Chiarugi, P. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res. 2010, 70, 6945–6956. [Google Scholar] [CrossRef] [Green Version]
- Honma, N.; Genda, T.; Matsuda, Y.; Yamagiwa, S.; Takamura, M.; Ichida, T.; Aoyagi, Y. MEK/ERK signaling is a critical mediator for integrin-induced cell scattering in highly metastatic hepatocellular carcinoma cells. Lab. Investig. 2006, 86, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Cowley, S.; Paterson, H.; Kemp, P.; Marshall, C.J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 1994, 77, 841–852. [Google Scholar] [CrossRef]
- Mansour, S.J.; Matten, W.T.; Hermann, A.S.; Candia, J.M.; Rong, S.; Fukasawa, K.; Vande Woude, G.F.; Ahn, N.G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 1994, 265, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Montesano, R.; Soriano, J.V.; Hosseini, G.; Pepper, M.S.; Schramek, H. Constitutively active mitogen-activated protein kinase kinase MEK1 disrupts morphogenesis and induces an invasive phenotype in Madin-Darby canine kidney epithelial cells. Cell Growth Differ. 1999, 10, 317–332. [Google Scholar] [PubMed]
- Pinkas, J.; Leder, P. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 2002, 62, 4781–4790. [Google Scholar]
- Voisin, L.; Julien, C.; Duhamel, S.; Gopalbhai, K.; Claveau, I.; Saba-El-Leil, M.K.; Rodrigue-Gervais, I.G.; Gaboury, L.; Lamarre, D.; Basik, M.; et al. Activation of MEK1 or MEK2 isoform is sufficient to fully transform intestinal epithelial cells and induce the formation of metastatic tumors. BMC Cancer 2008, 17, 337. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shen, M.X.; Ma, D.Z.; Wang, L.W.; Zha, X.L. TGF-β1-promoted epithelial- to–mesenchymal transformation and cell adhesion contribute to TGF-β 1-enhanced cell migration in SMMC-7721 cells. Cell Res. 2003, 13, 343–350. [Google Scholar] [CrossRef]
- Nantajit, D.; Lin, D.; Li, J.J. The network of epithelial-mesenchymal transition: Potential new targets for tumor resistance. J. Cancer Res. Clin. Oncol. 2015, 141, 1697–1713. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-β -induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Zhang, Y.E. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009, 19, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Attisano, L.; Wrana, J.L. Signal integration in TGF-β, WNT, and Hippo pathways. F1000Prime Rep. 2013, 5, 172013. [Google Scholar] [CrossRef]
- Morris, S.M.; Carter, K.T.; Baek, J.Y.; Koszarek, A.; Yeh, M.M.; Knoblaugh, S.E.; Grady, W.M. TGF-β signaling alters the pattern of liver tumorigenesis induced by Pten inactivation. Oncogene 2015, 34, 3273–3282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, Y.; Suzuki, A.; Kataoka, E.; Sasaki, T.; Hamada, K.; Sasaki, J.; Mizuno, K.; Hasegawa, G.; Kishimoto, H.; Iizuka, M.; et al. Hepatocyte-Specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Investig. 2004, 113, 1774–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikushima, H.; Miyazono, K. TGFβ signalling: A complex web in cancer progression. Nat. Rev. Cancer 2010, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Akhurst, R.J. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nat. Cell Biol. 2007, 9, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Shuang, L.; Chen, S.; Zeng, J. TGF-β signaling: A complex role in tumorigenesis. Mol. Med. Rep. 2018, 17, 699–704. [Google Scholar]
- Cordenonsi, M.; Dupont, S.; Maretto, S.; Insinga, A.; Imbriano, C.; Piccolo, S. Links between tumor suppressors: p53 is required for TGF-β gene responses by cooperating with Smads. Cell 2003, 113, 301–314. [Google Scholar] [CrossRef]
- Kalo, E.; Buganim, Y.; Shapira, K.E.; Besserglick, H.; Goldfinger, N.; Weisz, L.; Stambolsky, P.; Henis, Y.I.; Rotter, V. Mutant p53 attenuates the Smad-dependent transforming growth factor β1 (TGF-β1) signaling pathway by repressing the expression of TGF-β receptor type II. Mol. Cell. Biol. 2007, 27, 8228–8242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, Y.; Liu, Y.; Wang, N.; Qi, Y.; Du, J. Krüppel-Like factor 4 transcriptionally regulates TGF-β1 and contributes to cardiac myofibroblast differentiation. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Peng, Z.; Tang, H.; Xie, D.; Jia, Z.; Zhong, L.; Zhao, S.; Ma, Z.; Gao, Y.; Zeng, L.; et al. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-β signaling in and promote progression of hepatocellular carcinoma. Oncotarget 2017, 36, 2957–2968. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Zheng, B.; Zhang, Y.; Zhang, X.H.; Wang, C.; Yang, Z.; Sun, Y.; Wu, X.L.; Wen, J.K. KLF4 mediates the link between TGF-β1-induced gene transcription and H3 acetylation in vascular smooth muscle cells. FASEB J. 2015, 29, 4059–4070. [Google Scholar] [CrossRef]
- Podolec, J.; Baran, J.; Siedlinski, M.; Urbanczyk, M.; Krupinski, M.; Bartus, K.; Niewiara, L.; Podolec, M.; Guzik, T.; Tomkiewicz-Pajak, L.; et al. Serum rantes, transforming growth factor-β1 and interleukin-6 levels correlate with cardiac muscle fibrosis in patients with aortic valve stenosis. J. Physiol. Pharmacol. 2018, 69. [Google Scholar] [CrossRef]
- Wei, D.; Gong, W.; Kanai, M.; Schlunk, C.; Wang, L.; Yao, J.C.; Wu, T.T.; Huang, S.; Xie, K. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005, 65, 2746–2754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, D.; Kanai, M.; Huang, S.; Xie, K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 2006, 1, 23–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.X.; Han, M.; Bernier, M.; Zheng, B.; Sun, S.G.; Su, M.; Zhang, R.; Fu, J.R.; Wen, J.K. Krüppel-Like factor 4 promotes differentiation by transforming growth factor-beta receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells. J. Biol. Chem. 2010, 285, 17846–17856. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Kim, S.H.; Lim, J.W.; Kim, H. Lycopene induces apoptosis by inhibiting nuclear translocation of β-catenin in gastric cancer cells. J. Physiol. Pharmacol. 2019, 70. [Google Scholar] [CrossRef]
- Hashimoto, I.; Nagata, T.; Sekine, S.; Moriyama, M.; Shibuya, K.; Hojo, S.; Matsui, K.; Yoshioka, I.; Okumura, T.; Hori, T.; et al. Prognostic significance of KLF4 expression in gastric cancer. Oncol. Lett. 2017, 13, 819–826. [Google Scholar] [CrossRef] [Green Version]
- Cordenonsi, M.; Montagner, M.; Adorno, M.; Zacchigna, L.; Martello, G.; Mamidi, A.; Soligo, S.; Dupont, S.; Piccolo, S. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science 2007, 315, 840–843. [Google Scholar] [CrossRef]
- Elston, R.; Inman, G.J. Crosstalk between p53 and TGF-β Signalling. J. Signal Transduct. 2012, 294097, 1–10. [Google Scholar] [CrossRef] [Green Version]
| Gene | Primer Sequence | Size of PCR Product (bp) | Annealing Temp. (°C) |
|---|---|---|---|
| 18S | Forward 5′-GTTGGTTTTGATCTGATAAATGC-3′ Reverse 5′-CATTAAATCAGTTATGGTTCCTTTG-3′ | 143 | 60 |
| Bax | Forward 5′-CTGCCAACCCACCCTGGTCT-3′ Reverse 5′-TGGCAGCTGACATGTTTTCTG-3′ | 195 | 55 |
| BCL2 | Forward 5′-ACTGAGTACCTGAACCGGCATC-3′ Reverse 5′-GGAGAAATCAAACAGAGGTCGC-3′ | 108 | 60 |
| c-Myc | Forward 5′-CCACACAGCCCACTGGTCCT-3′ Reverse 5′-GGCTGGAGCATTTGCGGTTGTT-3′ | 163 | 60 |
| Cyclin D1 | Forward 5′-TGCTTGGGAAGTTGTGTTGG-3′ Reverse 5′-AATGCCATCACGGTCCCTAC-3′ | 126 | 60 |
| c-Myc | Forward 5′-CCACACAGCCCACTGGTCCT-3′ Reverse 5′-GGCTGGAGCATTTGCGGTTGTT-3′ | 163 | 60 |
| Ki-67 | Forward 5′-AACCAGGACTTTGTGCTCTGTAA-3′ Reverse 5′-CTCTTTTGGCTTCCATTTCTTC-3′ | 209 | 60 |
| Klf4 | Forward 5′-TTCTCCACGTTCGCGTCCGG-3′ Reverse 5′-TCTCGCCAACGGTTAGTCGGGG-3′ | 80 | 60 |
| Oct4 | Forward 5′-GGAGGGATGGCATACTGTGGACCT-3′ Reverse 5′-TCCTGGGACTCCTCGGGACTAGG-3′ | 197 | 60 |
| p53 | Forward 5′-AGTGAAGGGACTAGCATTGTC-3′ Reverse 5′-GGATGCCCGTGCTGCCGAGGAG-3′ | 243 | 60 |
| Sox2 | Forward 5′-AGAACCCCAAGATGCACAAC-3′ Reverse 5′-CTCCGGGAAGCGTGTACTTA-3′ | 204 | 60 |
| TGFβ rec 2 | Forward 5′-TGTGGCAGAGCGCTTCAGT-3′ Reverse 5′-TGTTCAGGGAGCCGTCTTCT-3′ | 95 | 60 |
| TGFβ rec 1 | Forward 5′-GCAGACTGGACCAGCAATGAC-3′ Reverse 5′-CTGCAATCAGGATCACTGCAA-3′ | 118 | 60 |
| Hp 16S | Forward 5′-GTCAAGAGATCAGCCTATGTCC-3′ Reverse 5′-TGGCAATCAGCGTCAGGTAATG-3′ | 522 | 54 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krzysiek-Maczka, G.; Targosz, A.; Szczyrk, U.; Wrobel, T.; Strzalka, M.; Brzozowski, T.; Czyz, J.; Ptak-Belowska, A. Long-Term Helicobacter pylori Infection Switches Gastric Epithelium Reprogramming towards Cancer Stem Cell-Related Differentiation Program in Hp-Activated Gastric Fibroblast-TGFβ Dependent Manner. Microorganisms 2020, 8, 1519. https://doi.org/10.3390/microorganisms8101519
Krzysiek-Maczka G, Targosz A, Szczyrk U, Wrobel T, Strzalka M, Brzozowski T, Czyz J, Ptak-Belowska A. Long-Term Helicobacter pylori Infection Switches Gastric Epithelium Reprogramming towards Cancer Stem Cell-Related Differentiation Program in Hp-Activated Gastric Fibroblast-TGFβ Dependent Manner. Microorganisms. 2020; 8(10):1519. https://doi.org/10.3390/microorganisms8101519
Chicago/Turabian StyleKrzysiek-Maczka, Gracjana, Aneta Targosz, Urszula Szczyrk, Tomasz Wrobel, Malgorzata Strzalka, Tomasz Brzozowski, Jaroslaw Czyz, and Agata Ptak-Belowska. 2020. "Long-Term Helicobacter pylori Infection Switches Gastric Epithelium Reprogramming towards Cancer Stem Cell-Related Differentiation Program in Hp-Activated Gastric Fibroblast-TGFβ Dependent Manner" Microorganisms 8, no. 10: 1519. https://doi.org/10.3390/microorganisms8101519
APA StyleKrzysiek-Maczka, G., Targosz, A., Szczyrk, U., Wrobel, T., Strzalka, M., Brzozowski, T., Czyz, J., & Ptak-Belowska, A. (2020). Long-Term Helicobacter pylori Infection Switches Gastric Epithelium Reprogramming towards Cancer Stem Cell-Related Differentiation Program in Hp-Activated Gastric Fibroblast-TGFβ Dependent Manner. Microorganisms, 8(10), 1519. https://doi.org/10.3390/microorganisms8101519







