Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials Used for the Preparation of Sourdough
2.2. Spontaneous Rye Sourdough Preparation and Sampling
2.3. Culture Media and Microbiological Growth and Enumeration
2.4. Isolation, Atype Identification of Sourdough Lactic Acid Bacteria Strains
2.5. Phenotype Characterization of the Isolated Sourdough Lactic Acid Bacteria Strains
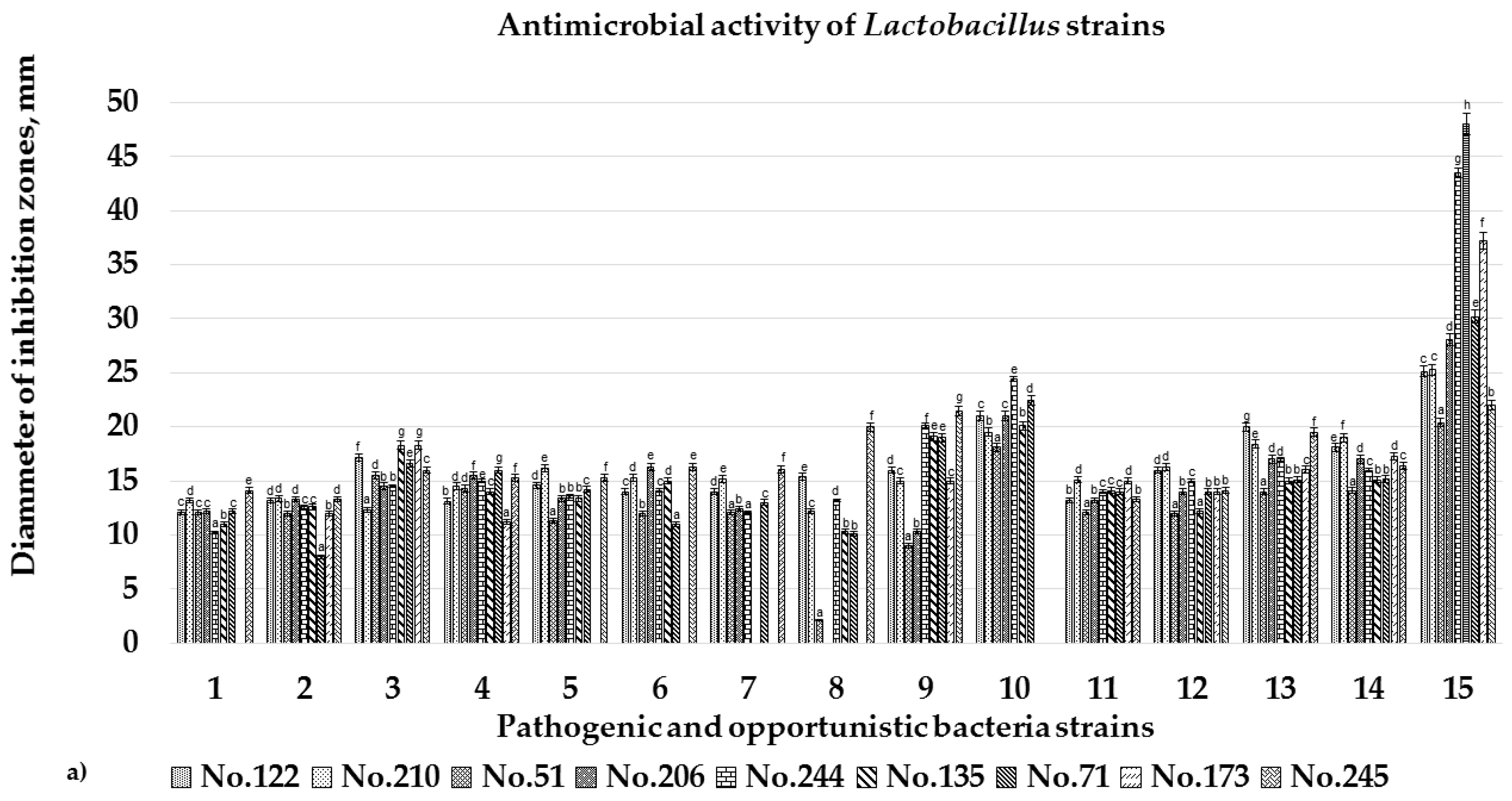
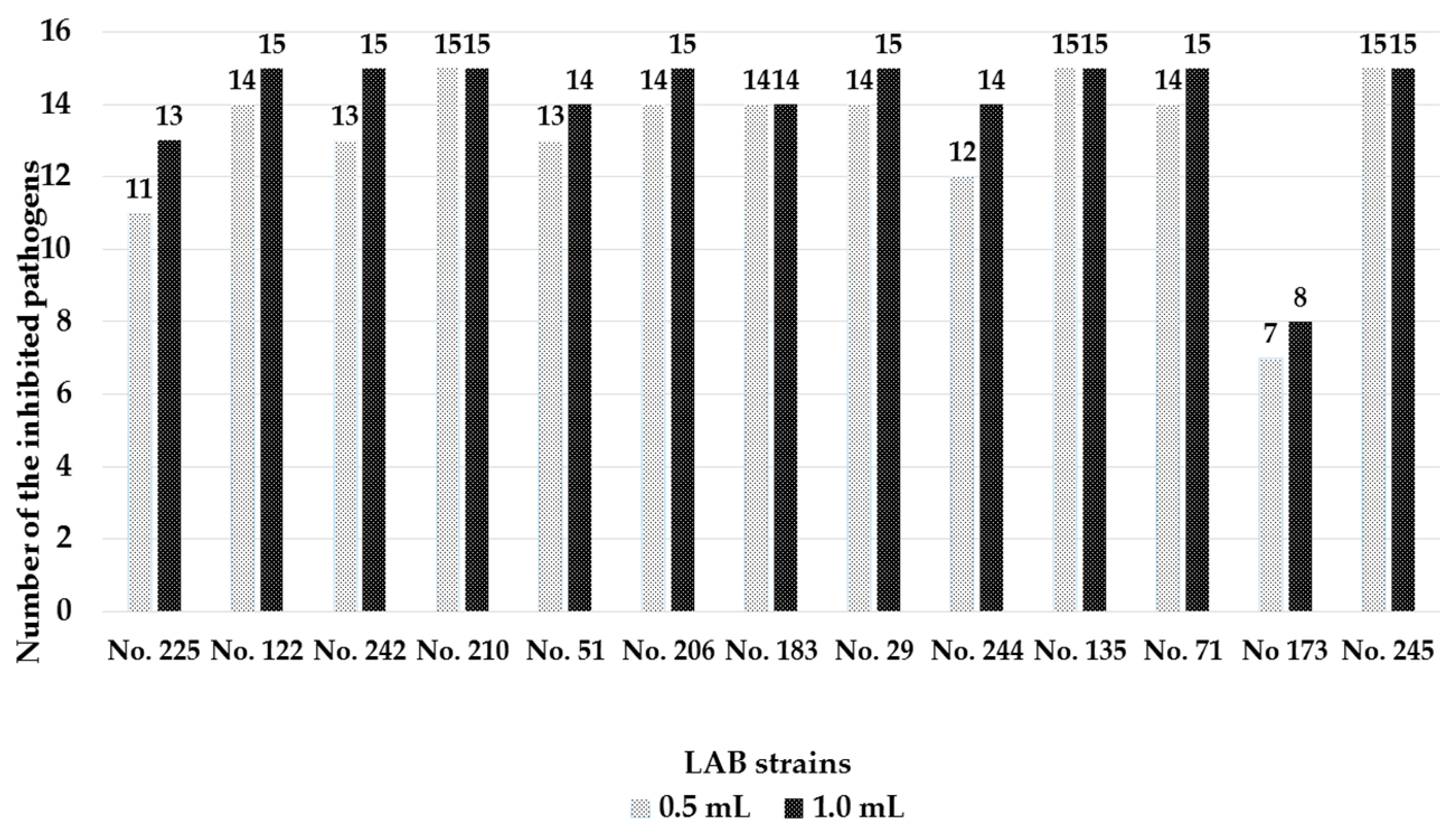
2.6. Antimicrobial Activity Testing of the Lactic Acid Bacteria Strains by Agar Well Diffusion Technique and Liquid Medium Based Methodology
2.7. Evaluation of Antifungal Activity of the Isolated Sourdough Lactic Acid Bacteria
2.8. Statistical Analysis
3. Results and Discussion
3.1. Genotype Identification of Lactic Acid Bacteria Strains Isolated from Spontaneous Rye Sourdough
3.2. Carbohydrate Metabolism, Gas Production, and Viability and Growth Performance at Different Temperatures and Low pH Values of Sourdough Isolates
3.3. Antimicrobial Activity of the Isolated Lactic Acid Bacteria Strains
3.4. Antifungal Activity of the Isolated Sourdough Lactic Acid Bacteria Strains
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rocha, J.M.; Malcata, F.X. Microbial Ecology Dynamics in Portuguese Broa Sourdough. J. Food Qual. 2016, 39, 634–648. [Google Scholar] [CrossRef]
- Rocha, J.M.; Malcata, F.X. Behavior of the Complex Micro-Ecology in Maize and Rye Flour and Mother-Dough for Broa throughout Storage. J. Food Qual. 2016, 39, 218–233. [Google Scholar] [CrossRef] [Green Version]
- Rocha, J.M.; Malcata, F.X. Microbiological profile of maize and rye flours, and sourdough used for the manufacture of traditional Portuguese bread. Food Microbiol. 2012, 31, 72–88. [Google Scholar] [CrossRef]
- Rocha, J.M. Microbiological and Lipid Profiles of Broa: Contributions for the Characterization of a Traditional Portuguese Bread; Superior School of Agriculture, University of Lisbon (ISA-UL): Lisbon, Portugal, 2011. [Google Scholar]
- Sakandar, H.A.; Hussain, R.; Kubow, S.; Sadiq, F.A.; Huang, W.; Imran, M. Sourdough bread: A contemporary cereal fermented product. J. Food Process. Preserv. 2019, 43, e13883. [Google Scholar] [CrossRef]
- Slavin, J. Whole grains and human health. Nutr. Res. Rev. 2004, 17, 99–110. [Google Scholar] [CrossRef] [Green Version]
- Catzeddu, P. Chapter 14—Sourdough Breads. In Flour and Breads and Their Fortification in Health and Disease Prevention, 2nd ed.; Preedy, V.R., Watson, R.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 177–188. ISBN 978-0-12-814639-2. [Google Scholar]
- Gobbetti, M.; Minervini, F.; Pontonio, E.; Di Cagno, R.; De Angelis, M. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. Int. J. Food Microbiol. 2016, 239, 3–18. [Google Scholar] [CrossRef]
- Zhang, G.; Tu, J.; Sadiq, F.A.; Zhang, W.; Wang, W. Prevalence, Genetic Diversity, and Technological Functions of the Lactobacillus sanfranciscensis in Sourdough: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1209–1226. [Google Scholar] [CrossRef] [Green Version]
- Rocha, J.M.; Malcata, F.X. On the microbiological profile of traditional Portuguese sourdough. J. Food Prot. 1999, 62, 1416–1429. [Google Scholar] [CrossRef] [PubMed]
- Sanpa, S.; Sanpa, S.; Suttajit, M. Lactic acid bacteria isolates from Pla-som, their antimicrobial activities and fermentation properties in Pla-som. J. Food Health Bioenvironmental Sci. 2019, 12, 36–43. [Google Scholar]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef]
- Aymerich, T.; Rodríguez, M.; Garriga, M.; Bover-Cid, S. Assessment of the bioprotective potential of lactic acid bacteria against Listeria monocytogenes on vacuum-packed cold-smoked salmon stored at 8 °C. Food Microbiol. 2019, 83, 64–70. [Google Scholar] [CrossRef] [PubMed]
- EFSA. ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- FDA. CFR—Code of Federal Regulations Title 21; 2019; Volume 21CFR184, p. 1538. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=184.1538 (accessed on 9 December 2019).
- Bartkiene, E.; Lele, V.; Sakiene, V.; Zavistanaviciute, P.; Ruzauskas, M.; Bernatoniene, J.; Jakstas, V.; Viskelis, P.; Zadeike, D.; Juodeikiene, G. Improvement of the antimicrobial activity of lactic acid bacteria in combination with berries/fruits and dairy industry by-products. J. Sci. Food Agric. 2019, 99, 3992–4002. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Bartkevics, V.; Mozuriene, E.; Krungleviciute, V.; Novoslavskij, A.; Santini, A.; Rozentale, I.; Juodeikiene, G.; Cizeikiene, D. The impact of lactic acid bacteria with antimicrobial properties on biodegradation of polycyclic aromatic hydrocarbons and biogenic amines in cold smoked pork sausages. Food Control 2017, 71, 285–292. [Google Scholar] [CrossRef]
- Bartkiene, E.; Ruzauskas, M.; Lele, V.; Zavistanaviciute, P.; Bernatoniene, J.; Jakstas, V.; Ivanauskas, L.; Zadeike, D.; Klupsaite, D.; Viskelis, P.; et al. Development of antimicrobial gummy candies with addition of bovine colostrum, essential oils and probiotics. Int. J. Food Sci. Technol. 2018, 53, 1227–1235. [Google Scholar] [CrossRef]
- Lele, V.; Ruzauskas, M.; Zavistanaviciute, P.; Laurusiene, R.; Rimene, G.; Kiudulaite, D.; Tomkeviciute, J.; Nemeikstyte, J.; Stankevicius, R.; Bartkiene, E. Development and characterization of the gummy–supplements, enriched with probiotics and prebiotics. CyTA J. Food 2018, 16, 580–587. [Google Scholar] [CrossRef]
- Bartkiene, E.; Bartkevics, V.; Krungleviciute, V.; Pugajeva, I.; Zadeike, D.; Juodeikiene, G. Lactic Acid Bacteria Combinations for Wheat Sourdough Preparation and Their Influence on Wheat Bread Quality and Acrylamide Formation. J. Food Sci. 2017, 82, 2371–2378. [Google Scholar] [CrossRef]
- Bartkiene, E.; Bartkevics, V.; Pugajeva, I.; Krungleviciute, V.; Mayrhofer, S.; Domig, K. The contribution of P. acidilactici, L. plantarum, and L. curvatus starters and L-(+)-lactic acid to the acrylamide content and quality parameters of mixed rye—Wheat bread. LWT 2017, 80, 43–50. [Google Scholar] [CrossRef]
- Bartkiene, E.; Bartkevics, V.; Krungleviciute, V.; Pugajeva, I.; Zadeike, D.; Juodeikiene, G.; Cizeikiene, D. The Influence of Scalded Flour, Fermentation, and Plants Belonging to Lamiaceae Family on the Wheat Bread Quality and Acrylamide Content. J. Food Sci. 2018, 83, 1560–1568. [Google Scholar] [CrossRef]
- Bartkiene, E.; Bartkevics, V.; Lele, V.; Pugajeva, I.; Zavistanaviciute, P.; Zadeike, D.; Juodeikiene, G. Application of antifungal lactobacilli in combination with coatings based on apple processing by-products as a bio-preservative in wheat bread production. J. Food Sci. Technol. 2019, 56, 2989–3000. [Google Scholar] [CrossRef]
- Krungleviciute, V.; Zelvyte, R.; Monkeviciene, I.; Kantautaite, J.; Stankevicius, R.; Ruzauskas, M.; Bartkiene, E. Applicability of Pediococcus strains for fermentation of cereal bran and its influence on the milk yield of dairy cattle. Zemdirb. Agric. 2017, 104, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Bartkiene, E.; Bartkevics, V.; Krungleviciute, V.; Juodeikiene, G.; Zadeike, D.; Baliukoniene, V.; Bakutis, B.; Zelvyte, R.; Santini, A.; Cizeikiene, D. Application of hydrolases and probiotic Pediococcus acidilactici BaltBio01 strain for cereal by-products conversion to bioproduct for food/feed. Int. J. Food Sci. Nutr. 2018, 69, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Zavistanaviciute, P.; Poskiene, I.; Lele, V.; Antanaitis, R.; Kantautaite, J.; Bartkiene, E. The influence of the newly isolated Lactobacillus plantarum LUHS135 and Lactobacillus paracasei LUHS244 strains on blood and faeces parametersin endurance horses. Pol. J. Vet. Sci. 2019, 22, 513–521. [Google Scholar] [PubMed]
- Bartkiene, E.; Bartkevics, V.; Lele, V.; Pugajeva, I.; Zavistanaviciute, P.; Mickiene, R.; Zadeike, D.; Juodeikiene, G. A concept of mould spoilage prevention and acrylamide reduction in wheat bread: Application of lactobacilli in combination with a cranberry coating. Food Control 2018, 91, 284–293. [Google Scholar] [CrossRef]
- Di Cello, F.; Bevivino, A.; Chiarini, L.; Fani, R.; Paffetti, D.; Tabacchioni, S.; Dalmastri, C. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 1997, 63, 4485–4493. [Google Scholar] [PubMed]
- Song, Y.; Kato, N.; Liu, C.; Matsumiya, Y.; Kato, H.; Watanabe, K. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 2000, 187, 167–173. [Google Scholar] [CrossRef]
- Berthier, F.; Ehrlich, S.D. Genetic diversity within Lactobacillus sakei and Lactobacillus curvatus and design of PCR primers for its detection using randomly amplified polymorphic DNA. Int. J. Syst. Evol. Microbiol. 1999, 49, 997–1007. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Meylan, V.; Klaenhammer, T.R.; Zink, R. Analysis, characterization, and loci of the tuf genes in lactobacillus and bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 2003, 69, 6908–6922. [Google Scholar] [CrossRef] [Green Version]
- Berthier, F.; Ehrlich, S.D. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 1998, 161, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Settanni, L.; van Sinderen, D.; Rossi, J.; Corsetti, A. Rapid differentiation and in situ detection of 16 sourdough lactobacillus species by multiplex PCR. Appl. Environ. Microbiol. 2005, 71, 3049–3059. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Park, S.Y.; Kim, J. Multiplex PCR-based detection and identification of Leuconostoc species. FEMS Microbiol. Lett. 2000, 193, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfannebecker, J.; Fröhlich, J. Use of a species-specific multiplex PCR for the identification of pediococci. Int. J. Food Microbiol. 2008, 128, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Mora, D.; Fortina, M.G.; Parini, C.; Manachini, P.L. Identification of Pediococcus acidilactici and Pediococcus pentosaceus based on 16S rRNA and ldhD gene-targeted multiplex PCR analysis. FEMS Microbiol. Lett. 1997, 151, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Harley, J.P.; Prescott, L.M.M. Laboratory Exercises in Microbiology, 7th ed.; McGraw-Hill Higher Education: New York City, NY, USA, 2008; ISBN 978-0-07-299293-9. [Google Scholar]
- Lee, J.; Yun, H.S.; Cho, K.W.; Oh, S.; Kim, S.H.; Chun, T.; Kim, B.; Whang, K.Y. Evaluation of probiotic characteristics of newly isolated Lactobacillus spp.: Immune modulation and longevity. Int. J. Food Microbiol. 2011, 148, 80–86. [Google Scholar] [CrossRef]
- International Organization for Standardization. Microbiology of Food and Animal Feeding Stuffs: Horizontal Method for the Detection of Listeria Monocytogenes. In Detection Methods; ISO: Geneva, Switzerland, 1996. [Google Scholar]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: M07-A10, Approved Standard, 10th ed.; Clinical and Laboratory Standards Institute (Ed.) Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2015; ISBN 978-1-56238-987-1. [Google Scholar]
- Cizeikiene, D.; Juodeikiene, G.; Paskevicius, A.; Bartkiene, E. Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 2013, 31, 539–545. [Google Scholar] [CrossRef]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Fu, W.; Rao, H.; Tian, Y.; Xue, W. Bacterial composition in sourdoughs from different regions in China and the microbial potential to reduce wheat allergens. LWT 2020, 117, 108669. [Google Scholar] [CrossRef]
- Yan, B.; Sadiq, F.A.; Cai, Y.; Fan, D.; Chen, W.; Zhang, H.; Zhao, J. Microbial diversity in traditional type I sourdough and jiaozi and its influence on volatiles in Chinese steamed bread. LWT 2019, 101, 764–773. [Google Scholar] [CrossRef]
- Fujimoto, A.; Ito, K.; Narushima, N.; Miyamoto, T. Identification of lactic acid bacteria and yeasts, and characterization of food components of sourdoughs used in Japanese bakeries. J. Biosci. Bioeng. 2019, 127, 575–581. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, M.; Jiaxin, C.; Luo, Y.; Ye, F.; Jiao, S.; Hu, X.; Zhang, J.; Lü, X. Bacterial diversity in traditional sourdough from different regions in China. LWT 2018, 96, 251–259. [Google Scholar] [CrossRef]
- Liu, A.; Jia, Y.; Zhao, L.; Gao, Y.; Liu, G.; Chen, Y.; Zhao, G.; Xu, L.; Shen, L.; Liu, Y.; et al. Diversity of isolated lactic acid bacteria in Ya’an sourdoughs and evaluation of their exopolysaccharide production characteristics. LWT 2018, 95, 17–22. [Google Scholar] [CrossRef]
- Zhao, Z.; Mu, T.; Sun, H. Microbial characterization of five Chinese traditional sourdoughs by high-throughput sequencing and their impact on the quality of potato steamed bread. Food Chem. 2019, 274, 710–717. [Google Scholar] [CrossRef]
- Dentice Maidana, S.; Aristimuño Ficoseco, C.; Bassi, D.; Cocconcelli, P.S.; Puglisi, E.; Savoy, G.; Vignolo, G.; Fontana, C. Biodiversity and technological-functional potential of lactic acid bacteria isolated from spontaneously fermented chia sourdough. Int. J. Food Microbiol. 2020, 316, 108425. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Zavistanaviciute, P.; Lele, V.; Ruzauskas, M.; Bartkevics, V.; Bernatoniene, J.; Gallo, P.; Tenore, G.C.; Santini, A. Lactobacillus plantarum LUHS135 and paracasei LUHS244 as functional starter cultures for the food fermentation industry: Characterisation, mycotoxin-reducing properties, optimisation of biomass growth and sustainable encapsulation by using dairy by-products. LWT 2018, 93, 649–658. [Google Scholar]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Guo, X.-H.; Kim, J.-M.; Nam, H.-M.; Park, S.-Y.; Kim, J.-M. Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 2010, 16, 321–326. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Pratsinis, H.; Nebe-von-Caron, G.; Kletsas, D.; Tsakalidou, E. Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 2007, 73, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Martín-Galiano, A.J.; Overweg, K.; Ferrándiz, M.J.; Reuter, M.; Wells, J.M.; de la Campa, A.G. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 2005, 151, 3935–3946. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou, K.; Boutou, E.; Zoumpopoulou, G.; Tarantilis, P.A.; Polissiou, M.; Vorgias, C.E.; Tsakalidou, E. RNA Arbitrarily Primed PCR and Fourier Transform Infrared Spectroscopy Reveal Plasticity in the Acid Tolerance Response of Streptococcus macedonicus. Appl. Environ. Microbiol. 2008, 74, 6068–6076. [Google Scholar] [CrossRef] [Green Version]
- Jayaraman, G.C.; Penders, J.E.; Burne, R.A. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 1997, 25, 329–341. [Google Scholar] [CrossRef]
- Wilkins, J.C.; Homer, K.A.; Beighton, D. Analysis of Streptococcus mutans Proteins Modulated by Culture under Acidic Conditions. Appl. Environ. Microbiol. 2002, 68, 2382–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaenhammer, T.R. Bacteriocins of lactic acid bacteria. Biochimie 1988, 70, 337–349. [Google Scholar] [CrossRef]
- Daeschel, M.A. Antimicrobial substances from lactic acid bacteria for use as food preservatives. Food Technol. 1989, 43, 164–167. [Google Scholar]
- Lindgren, S.E.; Dobrogosz, W.J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol. Rev. 1990, 7, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, U.; Lücke, F.K. Lactic acid bacteria as protective cultures in meat products. Fleischwirtschaft 1990, 70, 1296–1299. [Google Scholar]
- Piard, J.; Desmazeaud, M. Inhibiting factors produced by lactic acid bacteria. 1. Oxygen metabolites and catabolism end-products. Le Lait 1991, 71, 525–541. [Google Scholar] [CrossRef]
- Piard, J.; Desmazeaud, M. Inhibiting factors produced by lactic acid bacteria. 2. Bacteriocins and other antibacterial substances. Le Lait 1992, 72, 113–142. [Google Scholar] [CrossRef] [Green Version]
- Vandenbergh, P.A. Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol. Rev. 1993, 12, 221–237. [Google Scholar] [CrossRef]
- Li, J.; Xie, S.; Ahmed, S.; Wang, F.; Gu, Y.; Zhang, C.; Chai, X.; Wu, Y.; Cai, J.; Cheng, G. Antimicrobial Activity and Resistance: Influencing Factors. Front. Pharmacol. 2017, 8, 364. [Google Scholar] [CrossRef] [Green Version]
- Elayaraja, S.; Annamalai, N.; Mayavu, P.; Balasubramanian, T. Production, purification and characterization of bacteriocin from Lactobacillus murinus AU06 and its broad antibacterial spectrum. Asian Pac. J. Trop. Biomed. 2014, 4, S305–S311. [Google Scholar] [CrossRef] [Green Version]
- Fraberger, V.; Call, L.-M.; Domig, K.J.; D’Amico, S. Applicability of Yeast Fermentation to Reduce Fructans and Other FODMAPs. Nutrients 2018, 10, 1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, L.A.M.; Dal Bello, F.; Arendt, E.K. The use of sourdough fermented by antifungal LAB to reduce the amount of calcium propionate in bread. Int. J. Food Microbiol. 2008, 125, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut Microbiota: Role in Pathogen Colonization, Immune Responses and Inflammatory Disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Bartkiene, E.; Zokaityte, E.; Lele, V.; Sakiene, V.; Zavistanaviciute, P.; Klupsaite, D.; Bendoraitiene, J.; Navikaite-Snipaitiene, V.; Ruzauskas, M. Technology and characterisation of whole hemp seed beverages prepared from ultrasonicated and fermented whole seed paste. Int. J. Food Sci. Technol. 2019. [Google Scholar] [CrossRef]
- Rouse, S.; Harnett, D.; Vaughan, A.; van Sinderen, D. Lactic acid bacteria with potential to eliminate fungal spoilage in foods. J. Appl. Microbiol. 2008, 104, 915–923. [Google Scholar] [CrossRef]
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135. [Google Scholar] [CrossRef] [Green Version]

| Genus and Species | References | Primers Used (Fw = Forward; Re = Reverse) | Size (pb) |
|---|---|---|---|
| Lactobacillus | |||
| Lactobacillus spp. | [29] | Fw: 5′ CAA NTG GAT NGA ACC TGG CTT T3′ | 250 |
| Re: 5′ GCG TCA GGT TGG TGT TG3′ | |||
| Lactobacillus plantarum | [30] | Fw: 5′ GCT GGA TCA CCT CCT TTC 3′ | 248 |
| Re: 5′ ATG AGG TAT TCA ACT TAT G 3′ | |||
| Lactobacillus casei | [31] | Fw: 5′ CAA NTG GAT NGA ACC TGG CTT T 3′ | 520, 350 |
| Re: 5′ GAC GGT TAA GAT TGG TGA C 3′ | |||
| Lactobacillus paracasei | Fw: 5′ ACT GAA GGC GAC AAG GA 3′ | 520, 240 | |
| Re: 5′ GCG TCA GGT TGG TGT TG 3′ | |||
| Lactobacillus curvatus | [32] | Fw: 5′ GGA GGG TGT TCA GGA C 3′ | 260 |
| Re: 5′ GGA GGG TGT TGA TAG G 3′ | |||
| Lactobacillus brevis | [33] | Fw: 5′ GCC TTG SGA GAT GGT CCT C 3′ | 502 |
| Re: 5′TTT GAC GAT CAC GAA GTG ACC G 3′ | |||
| Leuconostoc | |||
| Leuconostoc mesenteroides | [34] | Fw: 5′ AAC TTA GTG TCG CAT GAC 3′ | 1150 |
| Re: 5′AGT CGA GTT ACA GAC TAC AA 3′ | |||
| Pediococcus | |||
| Pediococcus spp. | [35] | Fw: 5′ GAA CTC GTG TAC GTT GAA AAG TGC TGA 3′ | 701 |
| Re: 5′GCG TCC CTC CAT TGT TCA AAC AAG 3′ | |||
| Pediococcus pentosaceus | [36] | Fw: 5′ CGA ACT TCC GTT AAT TGA TCA G3′ | 872 |
| Re: 5′ACC TTG CGG TCG TAC TCC 3′ | |||
| Pediococcus acidilactici | Fw: 5′ CGA ACT TCC GTT AAT TGA TTA T3′ | 449 | |
| Re: 5′GTT CCG TCT TGC ATT TGA CC 3′ | |||
| 100bp DNA-Ladder Extended | Leuco-nostoc mesente-roides No. 242 | Lactoba-cillius corynifor-mins No. 71 | Lactoba-cillius curvatus No. 51 | Pediococcus pentosaceus No. 183 | Lactoba-cillius brevis No. 173 | Lactoba-cillius plantarum No. 135 |
 |  |  |  | 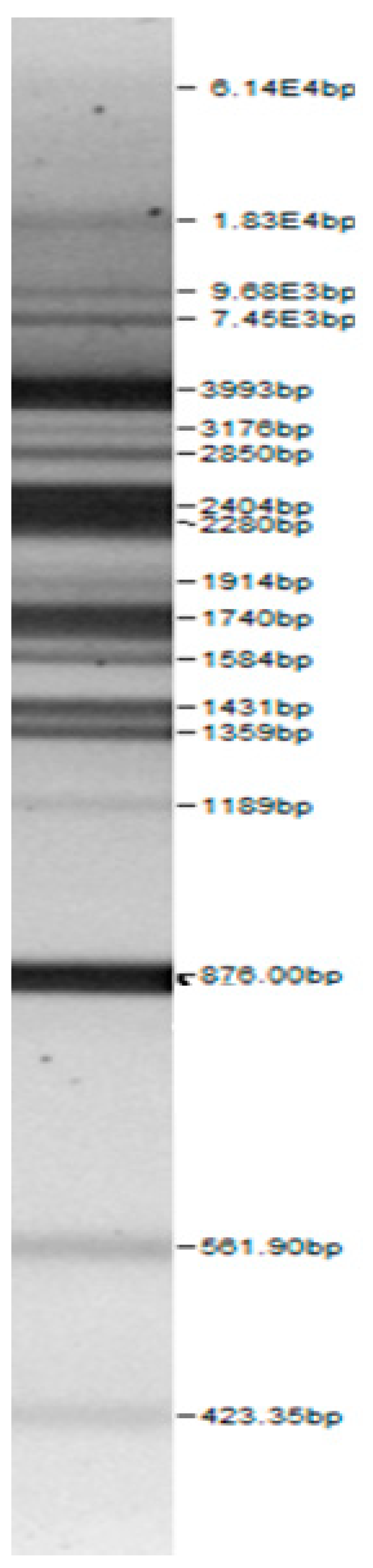 | 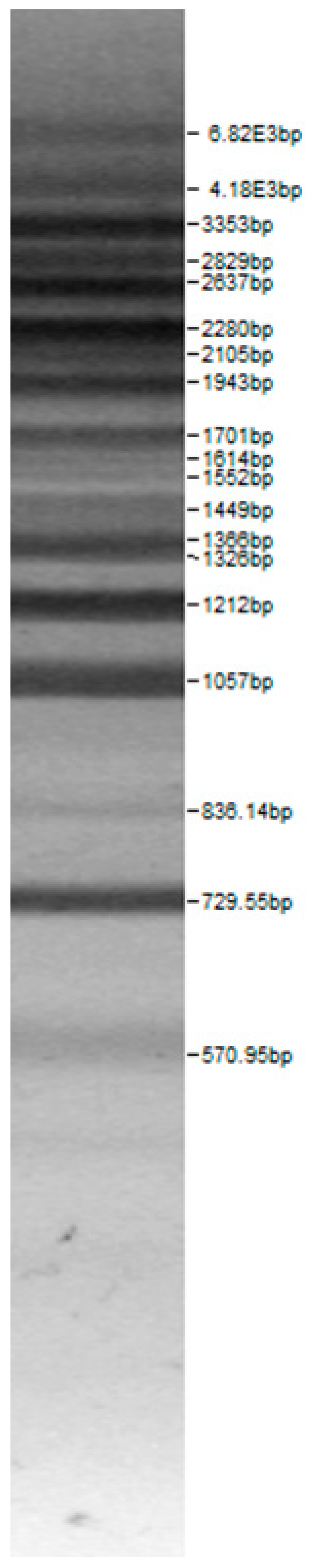 | 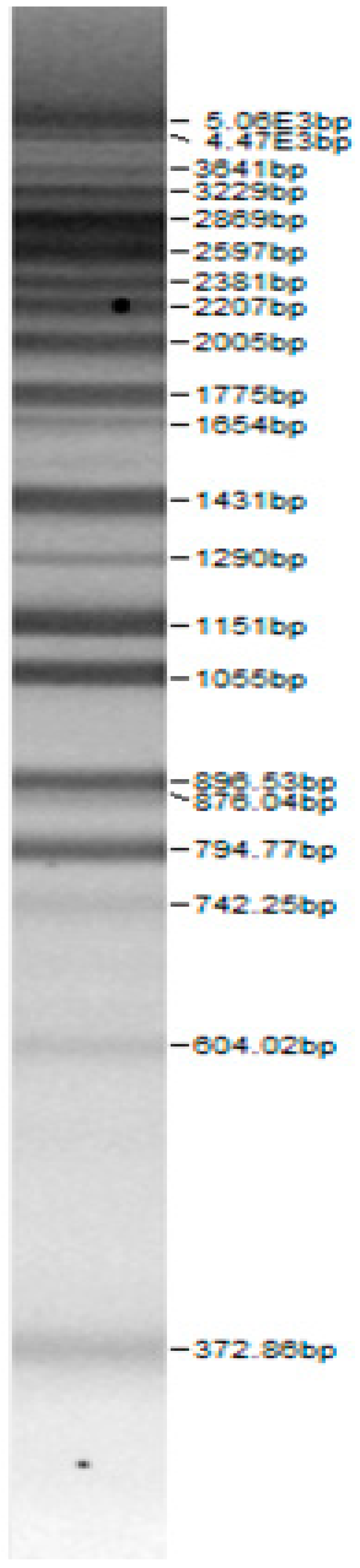 |
| Lactoba-cillius uvarum No. 245 | Lactoba-cillius plantarum No. 122 | Lactoba-cillius casei No. 210 | Leuconos-toc mesenteroi-des No. 225 | Lactobacillius farraginis No. 206 | Pedioco-ccus acidilac-tici No. 29 | Lactobaci-llius paracasei No. 244 |
 | 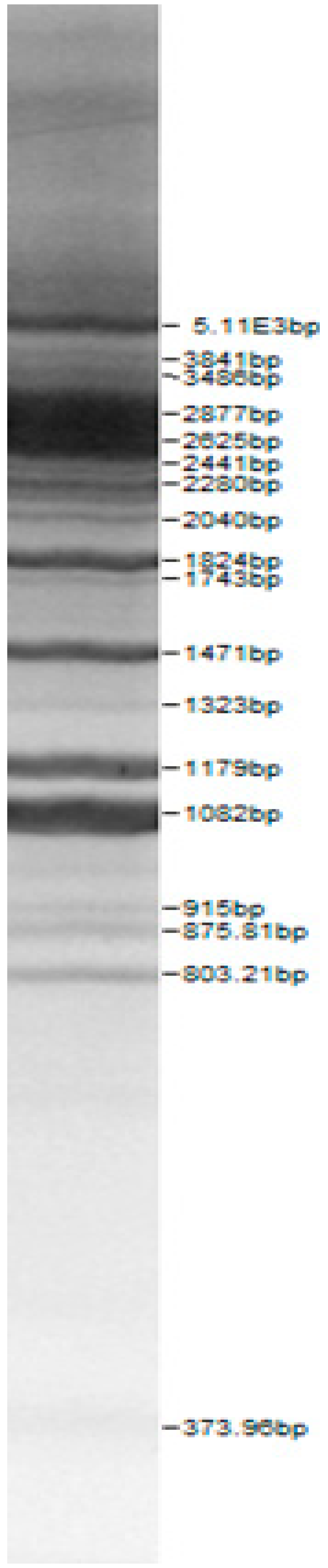 |  |  | 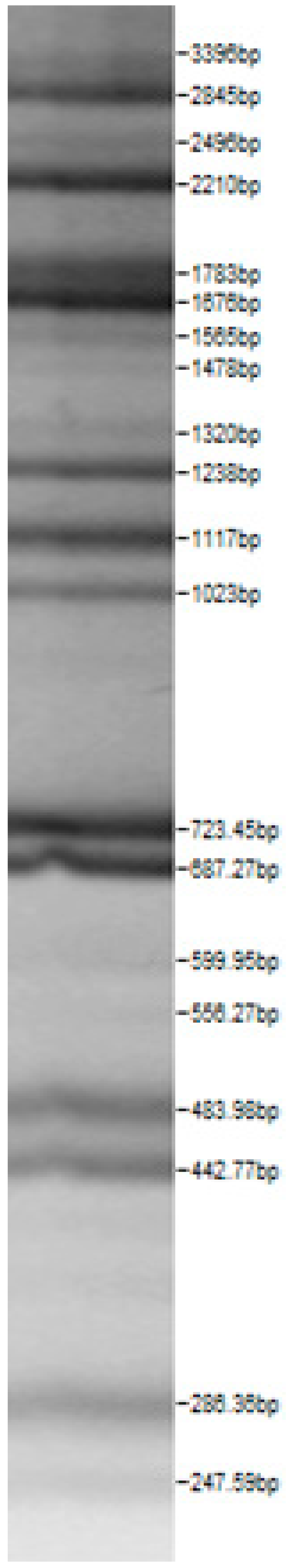 |  |  |
| Leu.mesenteroi-des No. 242 | L. coryniformins No. 71 | P. pentosaceus No. 183 | L. plantarum No. 122 | L. curvatus No. 51 | L. casei No. 210 | L. brevis No. 173 | L. uvarum No. 245 | Leu.mesenteroi-des No. 225 | L. farraginis No. 206 | L. plantarum No. 135 | P. acidilactici No. 29 | L. paracasei No. 244 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glicerol | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| d-arabinose | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| l-arabinose | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | − | |
| d-ribose | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | +++ | +++ | |
| d-xylose | +++ | − | − | − | − | − | +++ | − | − | +++ | − | +++ | − | |
| l-xylose | − | − | − | − | − | − | − | − | − | − | − | − | +++ | |
| d-adonitol | − | − | − | − | − | − | − | − | − | − | − | − | + | |
| Methyl-ßd-xYlopiranoside | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| d-galactose | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | + | +++ | +++ | +++ | |
| d-glucose | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| d-fructose | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |
| d-mannose | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| l-sorbose | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| l-rhamnose | − | + | ++ | + | − | − | − | − | − | − | + | ++ | +++ | |
| Dulcitol | − | − | − | − | − | +++ | − | − | − | − | − | − | +++ | |
| Inositol | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| d-mannitol | + | +++ | − | +++ | +++ | +++ | − | +++ | +++ | − | +++ | − | +++ | |
| d-sorbitol | − | +++ | − | +++ | +++ | +++ | − | +++ | − | − | +++ | − | +++ | |
| Methyl-αD-mannopyranoside | − | ++ | − | +++ | + | − | − | + | − | − | +++ | − | − | |
| Methyl-αD-glucopyranoside | +++ | − | − | − | − | +++ | − | − | +++ | − | + | − | +++ | |
| N-acetylglucosamine | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| Amigdalin | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| Arbutin | − | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| Esculin | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| Salicin | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| d-cellobiose | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| d-maltose | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | − | +++ | |
| d-lactose | − | +++ | − | +++ | +++ | − | − | +++ | − | − | +++ | +++ | +++ | |
| d-melibiose | +++ | ++ | +++ | +++ | − | − | − | − | − | +++ | +++ | − | − | |
| d-saccharose | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| d-trehalose | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | +++ | − | +++ | +++ | +++ | |
| Inulin | − | − | − | − | − | ++ | − | − | − | − | − | − | +++ | |
| d-melezitose | − | +++ | − | +++ | +++ | +++ | − | +++ | − | +++ | +++ | − | +++ | |
| d-raffinose | +++ | − | +++ | +++ | − | − | − | − | − | − | − | − | − | |
| Amidon | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Glycogen | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Xylitol | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Gentiobiose | ++ | +++ | +++ | ++ | ++ | ++ | − | ++ | +++ | − | ++ | +++ | +++ | |
| d-turanose | − | − | − | +++ | +++ | +++ | − | +++ | +++ | − | +++ | − | +++ | |
| d-tagatose | − | − | +++ | +++ | − | +++ | − | − | − | − | +++ | +++ | +++ | |
| d-fucose | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| l-fucose | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| d-arabitol | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| l-arabitol | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Potassium gluconate | + | − | − | + | + | + | + | + | − | ++ | ++ | + | ++ | |
| Potassium 2-ketogluconate | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| Potassium 5-ketogluconate | − | − | − | − | − | − | ++ | − | − | ++ | − | − | − | |
| Gas production (+/−) | + | − | − | − | − | − | + | − | − | − | − | − | − | |
| Temperature tolerance | 10 °C | − | − | − | − | − | + | − | − | − | − | − | − | − |
| 30 °C | + | +++ | ++ | ++ | + | +++ | + | ++ | ++ | + | ++ | ++ | ++ | |
| 37 °C | − | + | ++ | + | + | +++ | − | ++ | ++ | + | + | + | ++ | |
| 45 °C | − | − | − | + | − | + | − | − | − | ++ | + | + | − | |
| pH 2.5 | 0 h log (CFU/mL) | 8.14 ± 0.2 c | 6.51 ± 0.3 a | 7.97 ± 0.1 c | 8.43 ± 0.3 d | 8.31 ± 0.2 c,d | 8.47 ± 0.3 d | 8.86 ± 0.2 e | 9.03 ± 0.2 e | 8.14 ± 0.1c | 8.51 ± 0.2 d | 8.08 ± 0.2 c | 7.5 ± 0.2 b | 9.41 ± 0.2 f |
| 2 h log (CFU/mL) | 2.69 ± 0.1 a | n.d. | 7.40 ± 0.1 d | 5.72 ± 0.2 c | 3.5 ± 0.1 b | 8.36 ± 0.2 e | 8.67 ± 0.1 e | 7.55 ± 0.2 d | 2.69 ± 0.2 a | 8.42 ± 0.1e | 7.69 ± 0.1 d | 3.2 ± 0.1 b | 9.29 ± 0.1 f | |
| Isolated Sourdough Lactic Acid Bacteria (LAB) Strains | Aspergillus fischeri | Aspergillus nidulans | Penicillium oxalicum | Penicillium funiculosum | Fusarium poae | Alternaria alternata | Fusarium graminearum |
|---|---|---|---|---|---|---|---|
| Leuconostoc mesenteroides No. 225 | − | ++ | + | + | ++ | − | − |
| Lactobacillus plantarum No. 122 | − | ++ | +++ | +++ | +++ | ++ | ++ |
| Enteroccocus pseudoavium No. 242 | − | + | + | + | ++ | − | − |
| Lactobacillus casei No. 210 | − | ++ | + | ++ | +++ | + | + |
| Lactobacillus curvatus No. 51 | − | +++ | ++ | ++ | ++ | − | − |
| Lactobacillus farraginis No. 206 | + | + | ++ | + | +++ | + | − |
| Pediococcus pentosaceus No. 183 | − | ++ | ++ | +++ | + | + | − |
| Pediococcus acidilactici No. 29 | + | ++ | − | +++ | ++ | ++ | − |
| Lactobacillus paracasei No. 244 | + | + | + | +++ | +++ | − | + |
| Lactobacillus plantarum No. 135 | − | ++ | + | +++ | ++ | + | + |
| Lactobacillus coryniformins No. 71 | − | ++ | ++ | +++ | +++ | ++ | − |
| Lactobacillus brevis No. 173 | − | + | − | ++ | ++ | − | − |
| Lactobacillus uvarum No. 245 | − | ++ | + | +++ | + | − | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bartkiene, E.; Lele, V.; Ruzauskas, M.; Domig, K.J.; Starkute, V.; Zavistanaviciute, P.; Bartkevics, V.; Pugajeva, I.; Klupsaite, D.; Juodeikiene, G.; et al. Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms 2020, 8, 64. https://doi.org/10.3390/microorganisms8010064
Bartkiene E, Lele V, Ruzauskas M, Domig KJ, Starkute V, Zavistanaviciute P, Bartkevics V, Pugajeva I, Klupsaite D, Juodeikiene G, et al. Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms. 2020; 8(1):64. https://doi.org/10.3390/microorganisms8010064
Chicago/Turabian StyleBartkiene, Elena, Vita Lele, Modestas Ruzauskas, Konrad J. Domig, Vytaute Starkute, Paulina Zavistanaviciute, Vadims Bartkevics, Iveta Pugajeva, Dovile Klupsaite, Grazina Juodeikiene, and et al. 2020. "Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation" Microorganisms 8, no. 1: 64. https://doi.org/10.3390/microorganisms8010064
APA StyleBartkiene, E., Lele, V., Ruzauskas, M., Domig, K. J., Starkute, V., Zavistanaviciute, P., Bartkevics, V., Pugajeva, I., Klupsaite, D., Juodeikiene, G., Mickiene, R., & Rocha, J. M. (2020). Lactic Acid Bacteria Isolation from Spontaneous Sourdough and Their Characterization Including Antimicrobial and Antifungal Properties Evaluation. Microorganisms, 8(1), 64. https://doi.org/10.3390/microorganisms8010064










