Abstract
Cryptosporidium is an important protozoan parasite that can cause gastrointestinal diseases in humans and that also causes respiratory and gastrointestinal diseases in birds. In this study, we investigated the occurrence of Cryptosporidium species in migratory whooper swans in China. Fecal samples (n = 467) from whooper swans were collected from Sanmenxia Swan Lake National Urban Wetland Park, China. The samples were analyzed for Cryptosporidium species and genotypes with PCR along a sequence analysis of the small subunit rRNA. Cryptosporidium was detected in eight of the 467 (1.7%) samples. The analysis of the small subunit rRNA sequence data revealed two zoonotic species (Cryptosporidium parvum and Cryptosporidium andersoni) and one genotype (Cryptosporidium goose genotype II). These are the first data on the positive rate of Cryptosporidium spp. in whooper swans in China, and they suggest that whooper swans can harbor the zoonotic species C. parvum and C. andersoni in China.
1. Introduction
Cryptosporidium species are apicomplexan parasites and have a broad range of vertebrate hosts (including mammals and birds) [1,2]. The typical clinical symptom of Cryptosporidium infection in humans is watery diarrhea, which can lead to death in immunocompromised patients [3]. Symptoms such as diarrhea, cough, and dyspnea, as well as high morbidity and mortality may be observed in Cryptosporidium-infected birds [4], which can excrete feces containing Cryptosporidium oocysts into recreational and drinking water. Humans can acquire Cryptosporidium oocysts by ingesting contaminated food and water [5] or by direct contact with infected birds [6].
Based on molecular methods and morphological data, 38 Cryptosporidium species and more than 70 Cryptosporidium genotypes have been identified so far in various hosts [7]. Birds are mainly infected by Cryptosporidium baileyi, Cryptosporidium meleagridis, Cryptosporidium galli, and Cryptosporidium avium [8], but several other Cryptosporidium species and genotypes have been reported in birds in other studies, including Cryptosporidium parvum, Cryptosporidium andersoni, Cryptosporidium hominis, Cryptosporidium muris, Cryptosporidium goose genotypes (I–IV), a Cryptosporidium duck genotype, and Cryptosporidium avian genotypes (I–IV) [9,10].
The whooper swan (Cygnus cygnus) is a large, predominantly herbivorous, migratory waterfowl in the family Anatidae. It winters on freshwater lakes, brackish lagoons, coastal bays, low-lying coastal agricultural land and wet pastures. The whooper swan migrates along the middle and upper reaches of the Yellow River and its tributaries in spring after moving out of the Sanmenxia wetland, and it arrives at its breeding grounds in central and western Mongolia after flying over the Yinshan Mountains of China [11]. More than 10,000 whooper swans spend the winter in Sanmenxia Swan Lake National Urban Wetland Park every year, attracting many tourists. Tourists feed whooper swans in the Sanmenxia wetland, which is close to residential areas. Their excrement is scattered in public areas, including on grass and in water. Whooper swans reportedly migrate through at least 40 water bodies and lakes, posing a potential threat to public health [12].
Over 20 pathogenic microorganisms have been identified in swans to date, including avian influenza virus, Toxoplasma gondii, Microsporidia and Giardia [13,14,15,16,17]. However, the positive rate of Cryptosporidium and the number of Cryptosporidium species in whooper swans are unknown. Therefore, the aims of this study were to investigate the positive rate and species/genotypes of Cryptosporidium in whooper swans in China and to evaluate the potential threat of Cryptosporidium infection they pose to public health.
2. Materials and Methods
2.1. Ethics Approval
This study was conducted in accordance with the Law on the Protection of Wildlife adopted in 1988 in the People’s Republic of China. The research protocol was examined and approved by the Research Ethics Committee of Henan Agricultural University (Approval No. IRC-HENAU-20180919-05). Permission was obtained from all the managers of the study site before the fecal samples were collected.
2.2. Sample Collection
In total, 467 fecal specimens were collected from whooper swans in Sanmenxia Swan Lake National Urban Wetland Park in Sanmenxia city in the middle reaches of the Yellow River, China (Figure 1). In order to investigate the variation of the positive rate of Cryptosporidium in whooper swans over time and to further clarify the source of Cryptosporidium, we divided the sampling into three periods. Whooper swans arrive in the Sanmenxia wetland in November. Of the 467 samples, 237 were collected in November 2018, 161 were collected in December 2018, and 69 were collected in March 2019. The fresh fecal samples were collected immediately after defecation with a sterile disposal latex glove, and they were stored in 2.5% potassium dichromate at 4 °C before DNA extraction. To avoid the duplicated samples, each sample was collected in a short time with the help of the park staff.
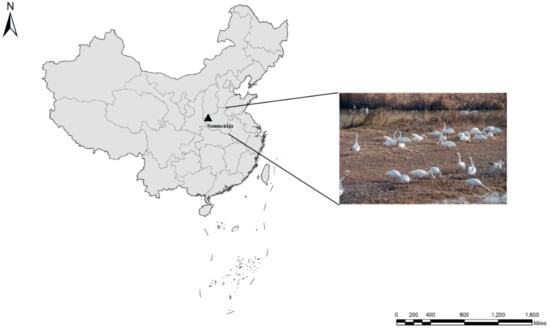
Figure 1.
Location of the city (▲) in which the samples were collected.
2.3. DNA Extraction
The genomic DNA was extracted from the fecal samples with the commercial E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA) according to the manufacturer’s instructions. The extracted DNA samples were stored at −20 °C before their PCR analysis for Cryptosporidium.
2.4. Cryptosporidium PCR Assays
Cryptosporidium species were detected with nested PCR targeting a fragment (approximately 830 bp) of the small subunit (SSU) rRNA gene. Xiao’s method, which was based on the small subunit rRNA sequence of Cryptosporidium, was used to design the nested PCR primers for the detection of Cryptosporidium [18,19]. The amplification was performed in 25 µL reaction mixtures. The first reaction mixture contained 1 μL of extracted DNA. The second reaction mixture contained 1 µL of the first PCR amplification product, and this was diluted 10-fold with sterile, double-distilled water. The KOD Plus DNA polymerase (Toyobo Co., Ltd., Osaka, Japan) was used for all PCR amplification. The second amplification products were examined with 1% (w/v) agarose gel electrophoresis after staining with DNA Green (Solarbio, Beijing, China). All PCRs were performed in triplicate, and both positive and negative control PCRs were included with each run.
2.5. Sequencing and Sequence Analysis
All positive amplification products were bidirectionally sequenced on an ABI PRISM™ 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) along with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). To determine the Cryptosporidium genotypes, ClustalX 2.1 (http://www.clustal.org/) was used to align each sequence with a reference sequence downloaded from GenBank. The phylogenetic relationships were determined with the neighbor-joining (NJ) algorithm in MEGA X (http://www.megasoftware.net/). On the phylogenetic tree, the lengths of the branches represent the evolutionary distances between genotypes, which were calculated according to the Kimura 2-parameter model. The accuracy of the tree was assessed with the bootstrap method with 1000 replicates.
3. Results
3.1. Cryptosporidium Positive Rate
According to the SSU-rRNA-based PCR, eight (1.7%) of the 467 fecal samples in this study were positive for Cryptosporidium. The positive rate of Cryptosporidium in the first sample group (2.9%, 7/237) was significantly higher than that in the second (0.62%, 1/161) and third sample groups (0%, 0/69; Table 1).

Table 1.
Cryptosporidium genotypes in whooper swans in China.
All PCR-positive samples were successfully sequenced. The nucleotide sequence analysis of the 18S rRNA revealed three distinct Cryptosporidium species or genotypes: C. parvum, C. andersoni, and Cryptosporidium goose genotype II (Table 1).
3.2. Phylogenetic Analysis of Cryptosporidium Isolates from Whooper Swans
An NJ tree was constructed from the 18S rRNA gene sequences that were isolated in this study and representative sequences from GenBank. The sequence from one fecal sample was identical to that of C. parvum (accession no. MN379944) and shared 100% identity with an isolate (AF093493) from a calf in the USA. C. andersoni (accession nos MN379937, MN379938, MN379940, MN379941, and MN379942) was detected in five specimens from whooper swans, with nucleotide sequences identical to the reference sequence (KJ094571) that was isolated from white yaks in China. The two remaining isolates were identical to that of Cryptosporidium goose genotype II (accession nos MN379939 and MN379943), and it shared 100% identity with an isolate from Canada geese (Branta canadensis) (AY504512) (Figure 2).
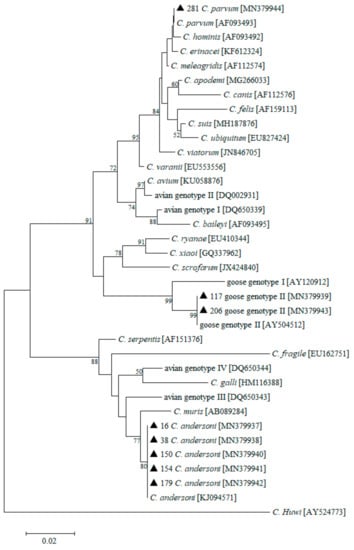
Figure 2.
Neighbor-joining tree based on small subunit (SSU) rRNA gene sequences of Cryptosporidium. GenBank accession numbers are shown in parentheses after the isolate identifiers. Numbers on the branches are percentage bootstrap values >50% that were calculated from 1000 replicates. Genotypes marked with filled triangles are known genotypes identified in the present study.
4. Discussion
This study is the first to investigate the positive rate and genotypes of Cryptosporidium spp. in whooper swans in China. The overall positive rate of Cryptosporidium was 1.7% (8/467) in these whooper swans, a rate which is similar to that previously reported in wild birds in Álava (2.3%, 6/265) and Algeria (2.8%, 2/71) [20,21]. However, in this study, the infection rate of Cryptosporidium was lower than those in free-living wild birds in Hungary (5.8% 6/103), Brazil (6.6% 16/242), and Spain (8.3% 36/433) [22,23,24]. Other studies have also reported much higher infection rates of Cryptosporidium in wild birds than this study, e.g., in wild captive psittacines in Brazil (10.64% 5/47), Java sparrows (Lonchura oryzivora) in northern China (13.42% 47/350), and North American red-winged blackbirds (Agelaius phoeniceus) (17.1% 12/70), and Canada geese (23.4% 49/209) in Ohio and Illinois [25,26,27,28]. Previous studies recorded Cryptosporidium infection rates in domestic birds of 2.3%–4.86% in Brazil [29,30,31] and 0.82%–8.1% in China [32,33,34,35,36]. These variations in the positive rate of Cryptosporidium in different studies may be attributable to population densities, feeding habits, and climate. Interestingly, the inconsistence in the positive rate of the three sample groups suggests that Cryptosporidium detected here might be from the origin or route of the migration instead of the sampling site.
Our study shows that two Cryptosporidium species and one genotype were identified in the whooper swans, namely C. parvum, C. andersoni, and Cryptosporidium goose genotype II. The zoonotic species C. parvum was detected in the whooper swans in this study, and this species has also been found in the feces of migratory Canada geese (Branta canadensis) in other studies [25,37]. C. parvum is responsible for most cases of cryptosporidiosis in humans and preweaned calves in China [38,39]. Previous reports have demonstrated that migratory herbivorous birds can acquire Cryptosporidium oocysts by ingesting undigested plant material in cattle feces. Other studies have reported that Cryptosporidium is found in wild birds, including the mute swan, white stork, carrion crow, mandarin duck, common merganser, rook, sparrowhawk, common buzzard, black kite, and honey buzzard [16,22]. C. parvum has been reported in domestic birds throughout the world, including Bengalese finches, psittacines, and cockatiels [31,40,41].
Recent studies have reported that C. andersoni can infect the quail-crested wood partridge [42], ostrich [43], and Canada goose [44]. However, there have been no reported cases of C. andersoni infection in domestic birds. It is well known that C. andersoni is the most common Cryptosporidium species in post-weaned calves and adult cattle, a fact which has been confirmed in China and Mongolia [45,46], and this species is known to infect humans [47].
In the present study, we detected Cryptosporidium goose genotype II in the feces of whooper swans. Natural infections of Cryptosporidium goose genotype II were first reported in Canada geese in America [25]. However, there have been no reports of infections of this genotype in other animals (including humans). Therefore, the public-health significance of this genotype remains unclear, and whether the whooper swan is a new host of this genotype still needs more investigation. Interestingly, four common species of Cryptosporidium in birds—C. baileyi, C. meleagridis, C. galli, and C. avium—were not identified in this survey. In addition, the dominant species/genotypes were different from each other in the three sample groups. Together with the above-discussed information, these data further indicate that the Cryptosporidium detected in the first sample groups might not originate from the sampling site but from the origin or route of the migration.
5. Conclusions
The presence of C. parvum, C. andersoni, and Cryptosporidium goose genotype II suggests that whooper swans may serve as transport hosts for Cryptosporidium spp. The inconsistence of species/genotypes detected in the three sample groups also suggests that whooper swans might play a role in the cross-regional transmission of Cryptosporidium, and this is the first report of Cryptosporidium in whooper swans.
Author Contributions
Conceptualization: L.Z.; methodology: L.Z.; formal analysis: K.W., A.G., Y.W., and K.Z.; investigation: K.W., A.G., Y.W., and K.Z.; data curation: K.W.; resources: Y.Z. (Yifan Zhang), Y.Z. (Yuxi Zhang), Y.C. (Yuan Cui); writing—original draft preparation: K.W.; writing—review and editing: Y.C. (Yankai Chang), S.Z. and L.Z.; project administration: L.Z.; funding acquisition: L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (grant numbers 2017YFD0500405 and 2017YFD0501305), the Natural Science Foundation of Henan Province (grant number 162300410129), and the Key Program of the National Natural Science Foundation of China (grant number 31330079).
Acknowledgments
We thank Janine Miller, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Ryan, U.; Xiao, R.F.L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology 2014, 141, 1667–1685. [Google Scholar] [CrossRef]
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef]
- Checkley, W.; White, A.C.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef]
- Santín, M. Clinical and subclinical infections with Cryptosporidium in animals. N. Z. Vet. J. 2013, 61, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Hijjawi, N.; Xiao, L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wesołowska, M.; Szostakowska, B.; Kicia, M.; Sak, B.; Kvac, M.; Knysz, B. Cryptosporidium meleagridis infection: The first report in Poland of its occurrence in an HIV-positive woman. Ann. Parasitol. 2016, 62, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Song, D.; Qi, M.; Zhang, S.; Wang, R.; Jian, F.; Ning, C.; Zhang, L. Revisiting the infectivity and pathogenicity of Cryptosporidium avium provides new information on parasitic sites within the host. Parasites Vectors 2018, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.A.; Meireles, M.V. Cryptosporidium infections in birds—A review. Rev. Bras. De Parasitol. Vet. 2015, 24, 253–267. [Google Scholar] [CrossRef]
- Ryan, U. Cryptosporidium in birds, fish and amphibians. Exp. Parasitol. 2010, 124, 113–120. [Google Scholar] [CrossRef]
- Li, S.; Meng, W.; Liu, D.; Yang, Q.; Chen, L.; Dai, Q.; Ma, T.; Gao, R.; Ru, W.; Li, Y.; et al. Migratory Whooper Swans Cygnus cygnus Transmit H5N1 Virus between China and Mongolia: Combination Evidence from Satellite Tracking and Phylogenetics Analysis. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, L.; Li, S.; Gao, R.; Ru, W.; Liu, D.; Sun, M.; Hou, Y.; Lu, J. The current status of wintering population of whooper swan (Cygnus cygnus) at Sanmenxia reservoir region, China. Chin. J. Zool. 2016, 51, 190–197. [Google Scholar] [CrossRef]
- Meng, W.; Yang, Q.; Vrancken, B.; Chen, Z.; Liu, D.; Chen, L.; Zhao, X.; Francois, S.; Ma, T.; Gao, R.; et al. New evidence for the east-west spread of the highly pathogenic avian influenza H5N1 virus between Central Asian and east Asian-Australasian flyways in China. Emerg. Microbes Infect. 2019, 8, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, H.; Lei, F.; Zhu, Q.; Qin, K.; Zhang, X.W.; Zhang, X.L.; Zhao, D.; Wang, G.; Feng, Y.; et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 2005, 309, 1206. [Google Scholar] [CrossRef] [PubMed]
- Slodkowicz-Kowalska, A.; Graczyk, T.K.; Tamang, L.; Jedrzejewski, S.; Nowosad, A.; Zduniak, P.; Solarczyk, P.; Girouard, A.S.; Majewska, A.C. Microsporidian species known to infect humans are present in aquatic birds: Implications for transmission via water? Appl. Environ. Microbiol. 2006, 72, 4540–4544. [Google Scholar] [CrossRef] [PubMed]
- Majewska, A.C.; Graczyk, T.K.; Slodkowicz-Kowalska, A.; Tamang, L.; Jedrzejewski, S.; Zduniak, P.; Solarczyk, P.; Nowosad, A.; Nowosad, P. The role of free-ranging, captive, and domestic birds of Western Poland in environmental contamination with Cryptosporidium parvum oocysts and Giardia lamblia cysts. Parasitol. Res. 2009, 104, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; Choudhary, S.; Kwok, O.C.; Ferreira, L.R.; Oliveira, S.; Verma, S.K.; Marks, D.R.; Pedersen, K.; Mickley, R.M.; Randall, A.R.; et al. Isolation and genetic characterization of Toxoplasma gondii from mute swan (Cygnus olor) from the USA. Vet. Parasitol. 2013, 195, 42–46. [Google Scholar] [CrossRef]
- Xiao, L.; Morgan, U.M.; Limor, J.; Escalante, A.; Arrowood, M.; Shulaw, W.; Thompson, R.C.; Fayer, R.; Lal, A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999, 65, 3386–3391. [Google Scholar] [PubMed]
- Xiao, L.; Singh, A.; Limor, J.; Graczyk, T.K.; Gradus, S.; Lal, A. Molecular characterization of cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 2001, 67, 1097–1101. [Google Scholar] [CrossRef]
- Cano, L.; de Lucio, A.; Bailo, B.; Cardona, G.A.; Muadica, A.S.; Lobo, L.; Carmena, D. Identification and genotyping of Giardia spp. and Cryptosporidium spp. isolates in aquatic birds in the Salburua wetlands, Alava, Northern Spain. Vet. Parasitol. 2016, 221, 144–148. [Google Scholar] [CrossRef]
- Elkarim Laatamna, A.; Holubova, N.; Sak, B.; Kvac, M. Cryptosporidium meleagridis and C. baileyi (Apicomplexa) in domestic and wild birds in Algeria. Folia Parasitol. 2017, 64. [Google Scholar] [CrossRef] [PubMed]
- Reboredo-Fernandez, A.; Ares-Mazas, E.; Caccio, S.M.; Gomez-Couso, H. Occurrence of Giardia and Cryptosporidium in wild birds in Galicia (Northwest Spain). Parasitology 2015, 142, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Sevá, A.d.P.; Funada, M.R.; Richtzenhain, L.; Guimarães, M.B.; Souza, S.D.O.; Allegretti, L.; Sinhorini, J.A.; Duarte, V.V.; Soares, R.M. Genotyping of Cryptosporidium spp. from free-living wild birds from Brazil. Vet. Parasitol. 2011, 175, 27–32. [Google Scholar] [CrossRef]
- Plutzer, J.; Tomor, B. The role of aquatic birds in the environmental dissemination of human pathogenic Giardia duodenalis cysts and Cryptosporidium oocysts in Hungary. Parasitol. Int. 2009, 58, 227–231. [Google Scholar] [CrossRef]
- Zhou, L.; Kassa, H.; Tischler, M.L.; Xiao, L. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Appl. Environ. Microbiol. 2004, 70, 4211–4215. [Google Scholar] [CrossRef][Green Version]
- Yao, Q.X.; Zhang, X.X.; Chen, K.; Ma, J.G.; Zheng, W.B.; Xu, X.Q.; Zhu, X.Q. Prevalence and Genetic Characterization of Cryptosporidium Infection in Java Sparrows (Lonchura oryzivora) in Northern China. BioMed Res. Int. 2017. [Google Scholar] [CrossRef]
- Oliveira, B.C.M.; Nagata, W.B.; Arana, D.G.; Ferreira, G.C.; Sitton, H.A.; de Oliveira, M.R.F.; Meireles, M.V. Cryptosporidium baileyi in wild captive psittacines in Brazil. Vet. Parasitol. Reg. Stud. Rep. 2017, 10, 154–156. [Google Scholar] [CrossRef]
- Chelladurai, J.J.; Clark, M.E.; Kvac, M.; Holubova, N.; Khan, E.; Stenger, B.L.; Giddings, C.W.; McEvoy, J. Cryptosporidium galli and novel Cryptosporidium avian genotype VI in North American red-winged blackbirds (Agelaius phoeniceus). Parasitol. Res. 2016, 115, 1901–1906. [Google Scholar] [CrossRef]
- Camargo, V.d.S.; Santana, B.N.; Ferrari, E.D.; Nakamura, A.A.; Nagata, W.B.; Nardi, A.R.M.; Meireles, M.V. Detection and molecular characterization of Cryptosporidium spp. in captive canaries (Serinus canaria) using different diagnostic methods. Rev. Bras. De Parasitol. Vet. 2018, 27, 60–65. [Google Scholar] [CrossRef]
- Da Cunha, M.J.R.; Cury, M.C.; Santin, M. Molecular identification of Enterocytozoon bieneusi, Cryptosporidium, and Giardia in Brazilian captive birds. Parasitol. Res. 2017, 116, 487–493. [Google Scholar] [CrossRef]
- Nakamura, A.A.; Simões, D.C.; Antunes, R.G.; da Silva, D.C.; Meireles, M.V. Molecular characterization of Cryptosporidium spp. from fecal samples of birds kept in captivity in Brazil. Vet. Parasitol. 2009, 166, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.-Y.; Chang, H.; Luo, J.; Huang, J.-J.; He, H.-X. First report of Enterocytozoon bieneusi and Cryptosporidium spp. in peafowl (Pavo cristatus) in China. Int. J. Parasitol. Parasites Wildl. 2019, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-X.; Zhang, N.-Z.; Zhao, G.-H.; Zhao, Q.; Zhu, X.-Q. Prevalence and Genotyping of Cryptosporidium Infection in Pet Parrots in North China. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.; Zhang, L.; Qi, N.; Liao, S.; Lv, M.; Wu, C.; Sun, M. Molecular characterization of Cryptosporidium spp. in domestic pigeons (Columba livia domestica) in Guangdong Province, Southern China. Parasitol. Res. 2015, 114, 2237–2241. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, R.; Ning, C.; Li, X.; Zhang, L.; Jian, F.; Sun, Y.; Xiao, L. Cryptosporidium spp. in pet birds: Genetic diversity and potential public health significance. Exp. Parasitol. 2011, 128, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Huang, L.; Wang, R.; Xiao, L.; Xu, L.; Li, J.; Zhang, L. Natural infection of Cryptosporidium muris in ostriches (Struthio camelus). Vet. Parasitol. 2014, 205, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, T.K.; Fayer, R.; Trout, J.M.; Lewis, E.J.; Farley, C.A.; Sulaiman, I.; Lal, A.A. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl. Environ. Microbiol. 1998, 64, 2736–2738. [Google Scholar]
- Qi, M.; Wang, H.; Jing, B.; Wang, D.; Wang, R.; Zhang, L. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol. 2015, 212, 404–407. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Zhao, X.; Zhang, L.; Zhang, G.; Guo, M.; Liu, L.; Feng, Y.; Xiao, L. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013, 51, 557–563. [Google Scholar] [CrossRef]
- Gomes, R.S.; Huber, F.; da Silva, S.; do Bomfim, T.C. Cryptosporidium spp. parasitize exotic birds that are commercialized in markets, commercial aviaries, and pet shops. Parasitol. Res. 2012, 110, 1363–1370. [Google Scholar] [CrossRef]
- Holubová, N.; Zikmundová, V.; Limpouchová, Z.; Sak, B.; Konečný, R.; Hlásková, L.; Rajský, D.; Kopacz, Z.; McEvoy, J.; Kváč, M. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur. J. Protistol. 2019, 69, 70–87. [Google Scholar] [CrossRef]
- Ng, J.; Pavlasek, I.; Ryan, U. Identification of novel Cryptosporidium genotypes from avian hosts. Appl. Environ. Microbiol. 2006, 72, 7548–7553. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.; El Safadi, D.; Benamrouz-Vanneste, S.; Cian, A.; Moriniere, R.; Gantois, N.; Delgado-Viscogliosi, P.; Guyot, K.; Bosc, S.; Chabe, M.; et al. Prevalence, transmission, and host specificity of Cryptosporidium spp. in various animal groups from two French zoos. Parasitol. Res. 2017, 116, 3419–3422. [Google Scholar] [CrossRef] [PubMed]
- Kassa, H.; Harrington, B.J.; Bisesi, M.S. Cryptosporidiosis: A brief literature review and update regarding Cryptosporidium in feces of Canada geese (Branta canadensis). J. Environ. Health 2004, 66, 34–40, 45. [Google Scholar] [PubMed]
- Liang, N.; Wu, Y.; Sun, M.; Chang, Y.; Lin, X.; Yu, L.; Hu, S.; Zhang, X.; Zheng, S.; Cui, Z.; et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology 2019, 146, 28–32. [Google Scholar] [CrossRef]
- Burenbaatar, B.; Bakheit, M.A.; Plutzer, J.; Suzuki, N.; Igarashi, I.; Ongerth, J.; Karanis, P. Prevalence and genotyping of Cryptosporidium species from farm animals in Mongolia. Parasitol. Res. 2008, 102, 901–905. [Google Scholar] [CrossRef]
- Liu, H.; Shen, Y.; Yin, J.; Yuan, Z.; Jiang, Y.; Xu, Y.; Pan, W.; Hu, Y.; Cao, J. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect. Dis. 2014, 14, 25. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).