Enrichment and Analysis of Stable 1,4-dioxane-Degrading Microbial Consortia Consisting of Novel Dioxane-Degraders
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
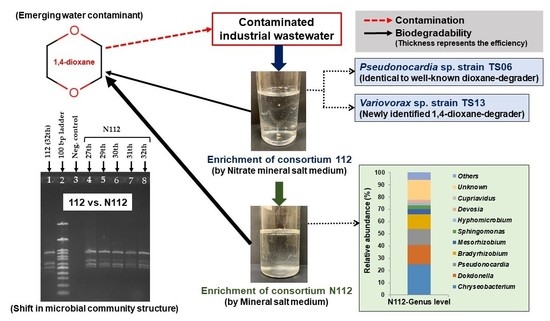
2.2. Enrichment of 1,4-dioxane-Degrading Microbial Consortia
2.3. Analytical Methods
2.4. Degradation Experiments
2.5. Isolation and Identification of 1,4-dioxane-Degrading Strains from Consortium 112
2.6. Microbial Community Analysis
2.7. Quantitative PCR (qPCR) Analysis
2.8. Accession Number
3. Results
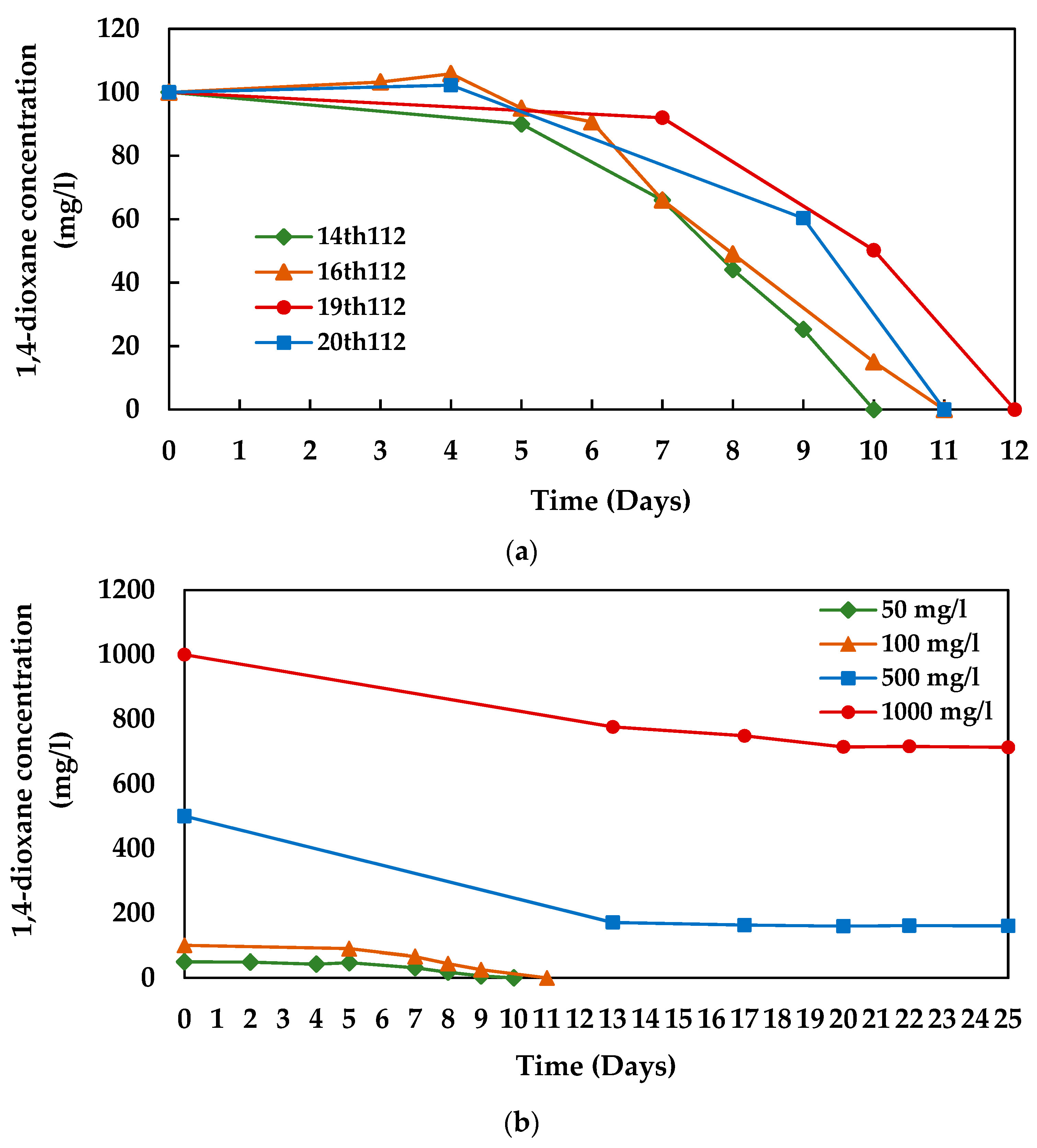
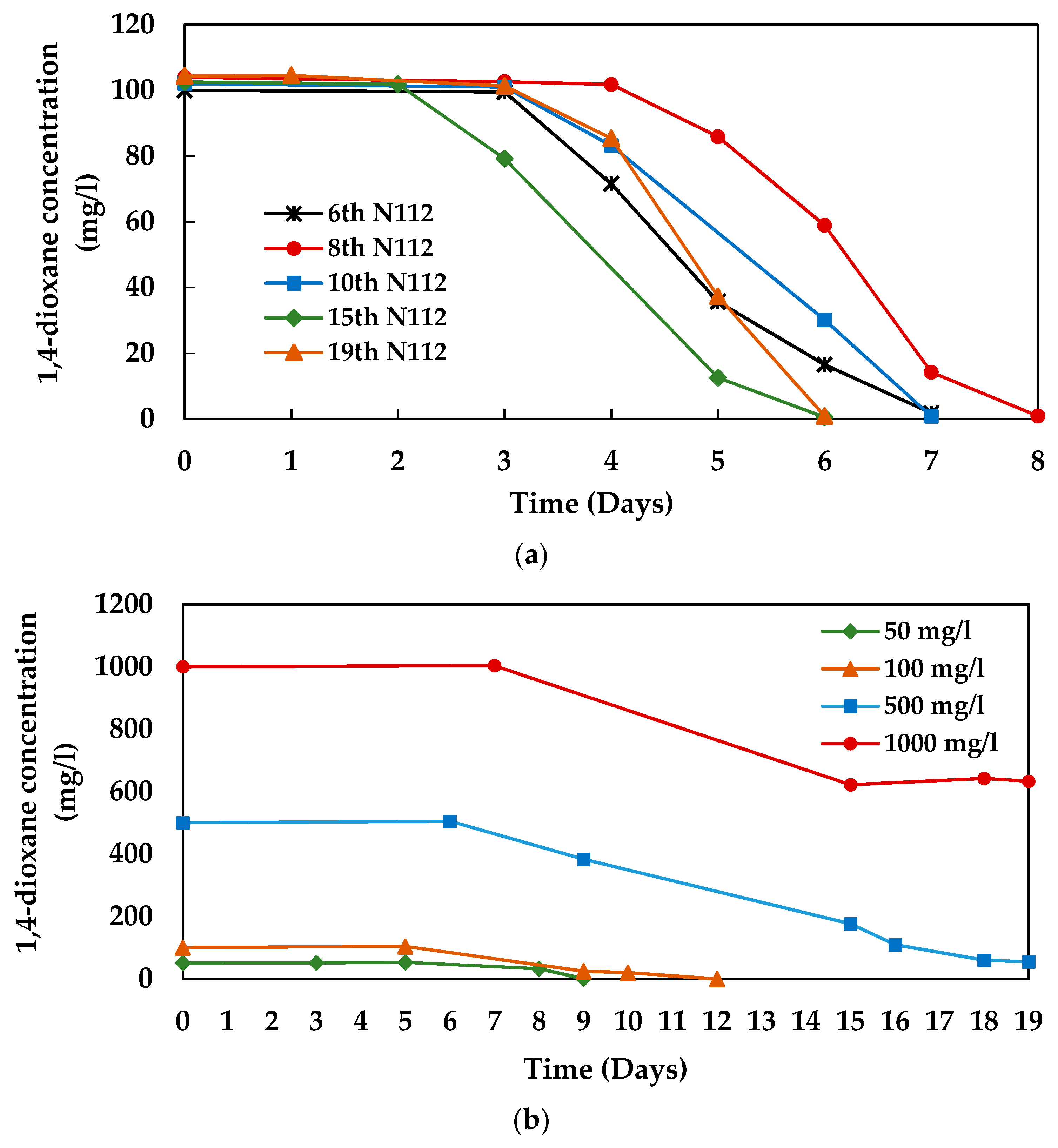
3.1. 1,4-dioxane Degradation Efficiency of Microbial Consortium 112
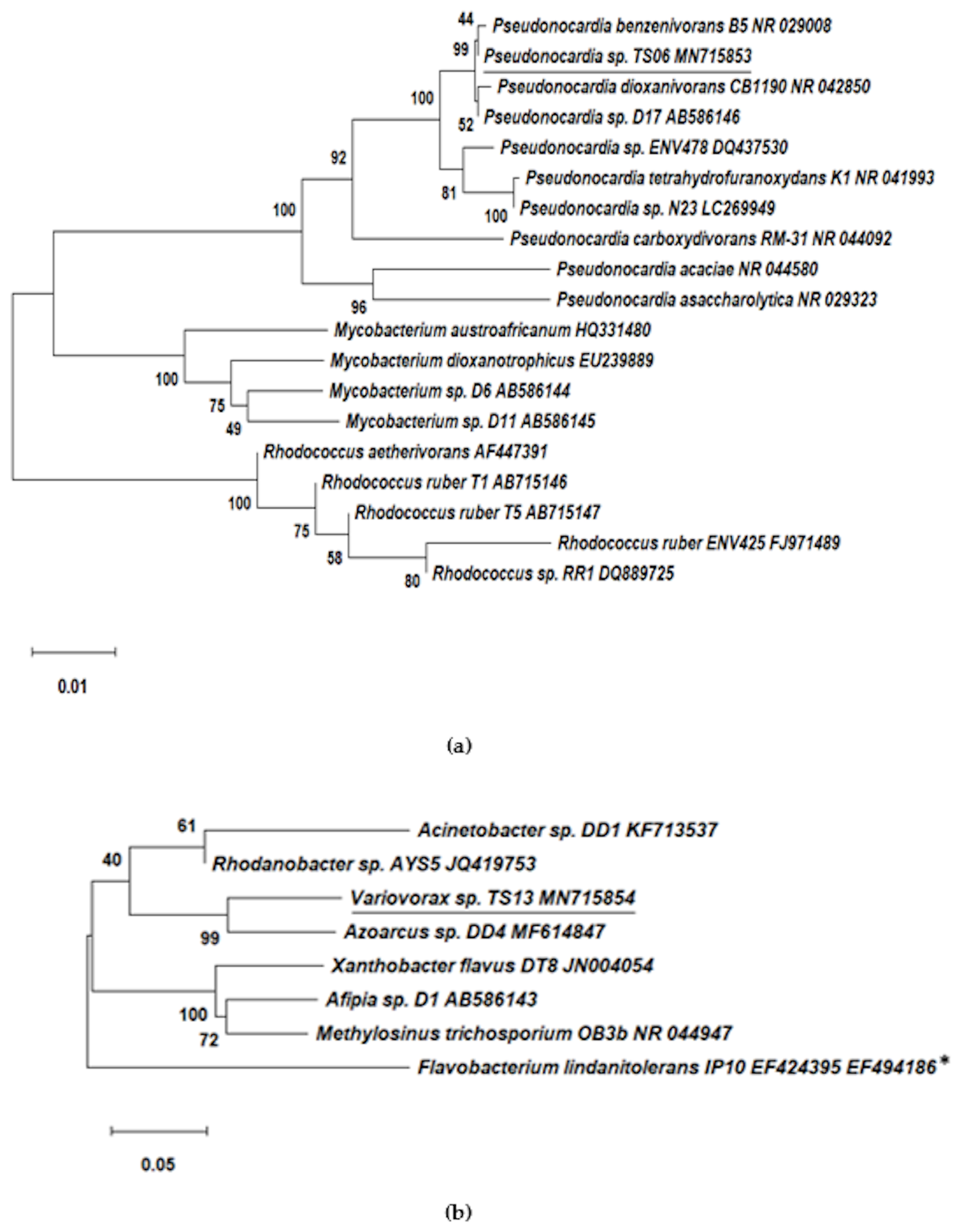
3.2. Isolation and Identification of 1,4-dioxane-Degrading Strains from Consortium 112
3.3. 1,4-dioxane Degradation Efficiency of Consortium N112
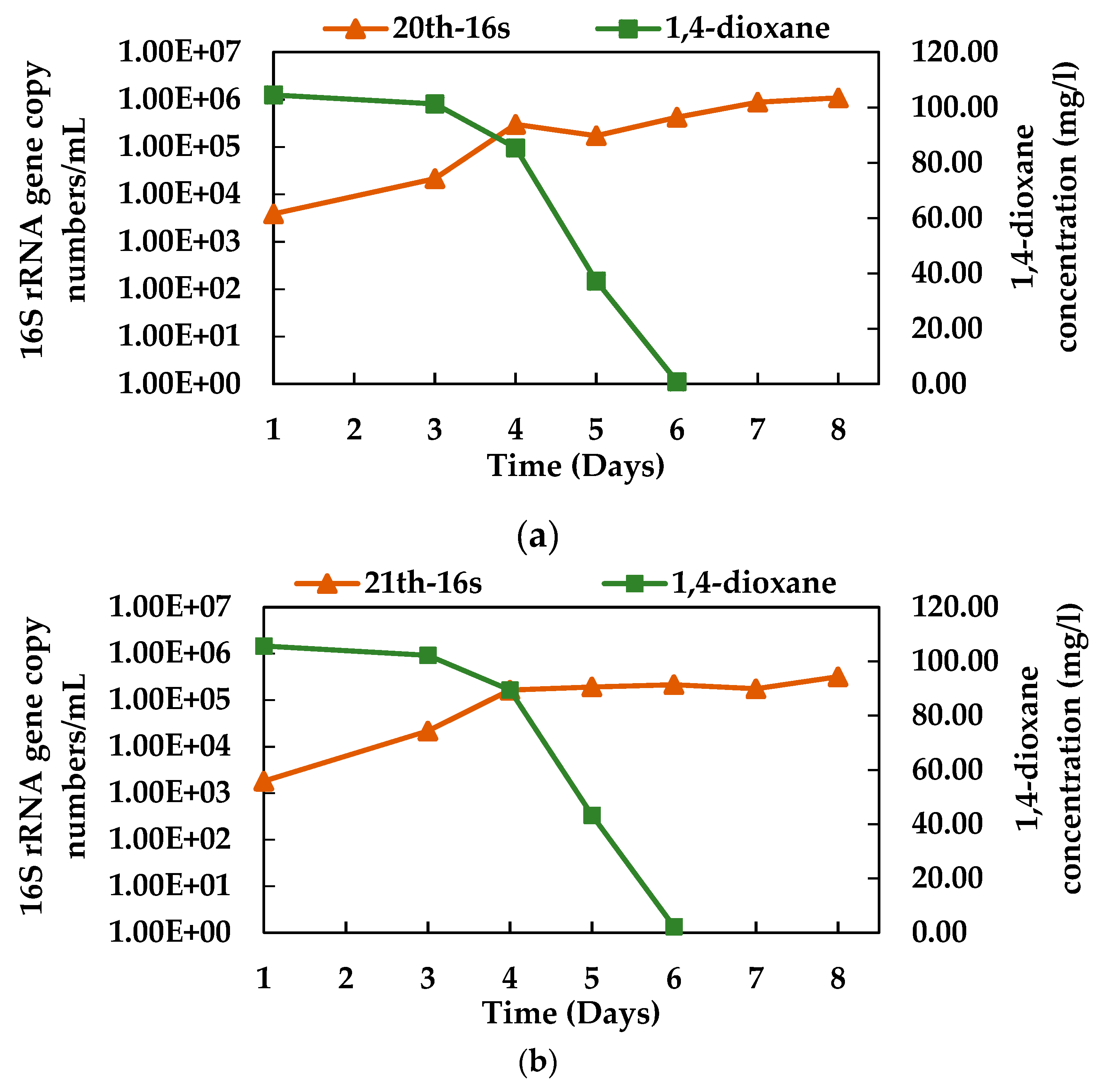
3.4. Growth of N112 with 1,4-dioxane Degradation
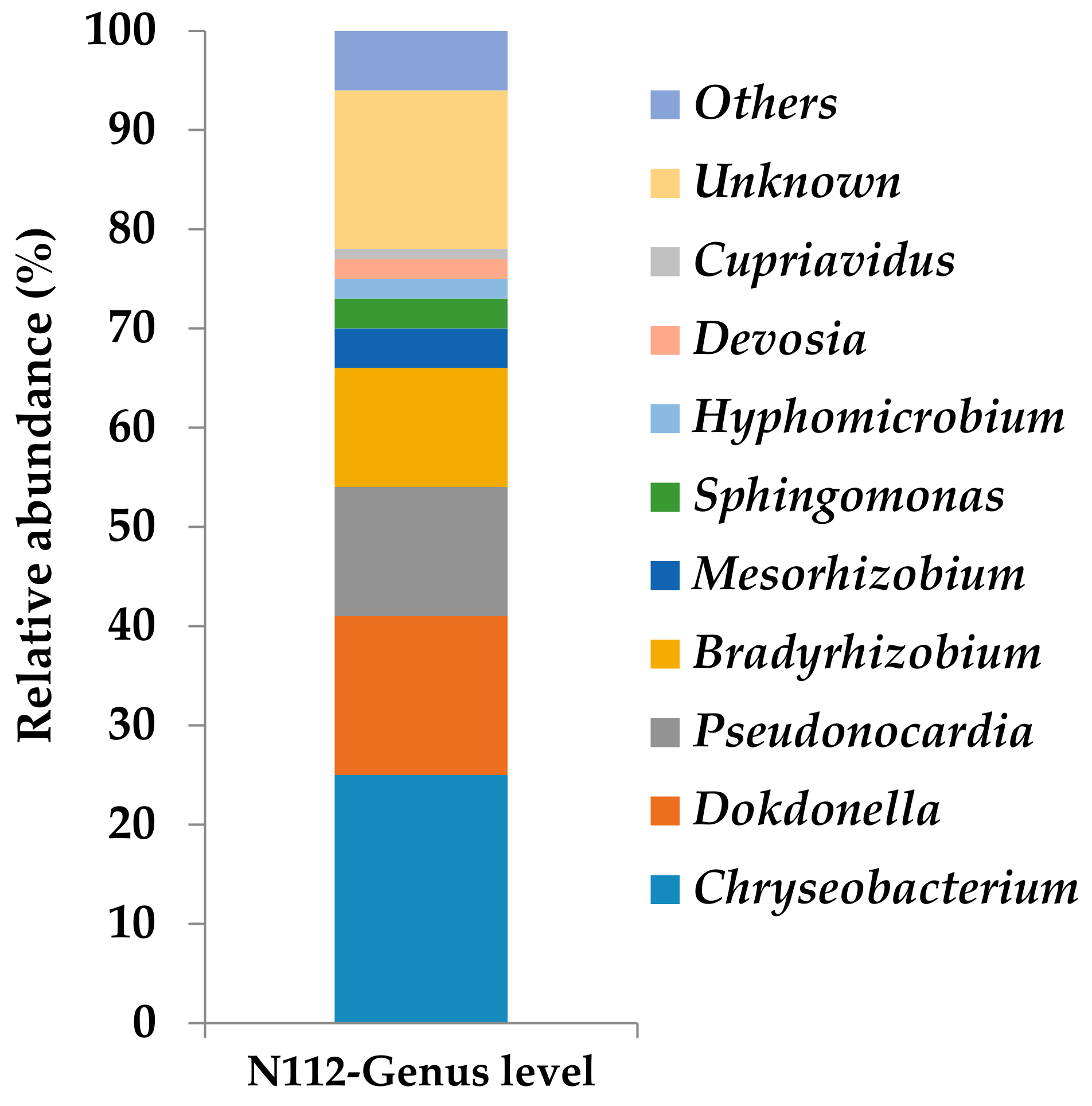
3.5. Analysis of the Microbial Community Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, D.Z.; Jin, X.J.; Chen, J.; Ye, J.X.; Jiang, N.X.; Chen, J.M. Intermediates and substrate interaction of 1,4-dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. Int. Biodeter. Biodegr. 2016, 106, 133–140. [Google Scholar] [CrossRef]
- Sei, K.; Miyagaki, K.; Kakinoki, T.; Fukugasako, K.; Inoue, D.; Ike, M. Isolation and characterization of bacterial strains that have high ability to degrade 1,4-dioxane as a sole carbon and energy source. Biodegradation 2013, 24, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gedalanga, P.B.; Mahendra, S. Advances in bioremediation of 1,4-dioxane-contaminated waters. J. Environ. Manag. 2013, 24, 665–674. [Google Scholar] [CrossRef] [PubMed]
- IARC (International Agency for Research on Cancer). Re-evaluation of some organic chemicals hydrazine and hydrogen peroxide. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; No. 71; IARC: Lyon, France, 1999. [Google Scholar]
- USEPA (U.S. Environmental Protection Agency). Toxicological review of 1,4-dioxane (CAS No. 123-91-1). In Integrated Risk Information System (IRIS); USEPA: Washington, DC, USA, 2013. [Google Scholar]
- Pollitt, K.J.G.; Kim, J.H.; Peccia, J.; Elimelech, M.; Zhang, Y.; Charkoftaki, G.; Hodges, B.; Zucker, I.; Huang, H.; Deziel, N.C.; et al. 1,4-dioxane as an emerging water contaminant: State of the science and evaluation of research fields. Sci. Total Environ. 2019, 690, 853–866. [Google Scholar] [CrossRef]
- ATSDR (Agency for Toxic Substances and Disease Registry). Support Document to the 2017 Substance Priority List (Candidates for Toxicological Profiles); Division of Toxicology and Human Health Sciences, ATSDR: Atlanta, GA, USA, 2017.
- Zenker, M.J.; Borden, R.C.; Barlaz, M.A. Occurrence and treatment of 1,4-dioxane in aqueous environments. Environ. Eng. Sci. 2003, 20, 423–432. [Google Scholar] [CrossRef]
- Inoue, D.; Tsunoda, T.; Sawada, K.; Yamamoto, N.; Saito, Y.; Sei, K.; Ike, M. 1,4-dioxane degradation potential of members of the genera Pseudonocardia and Rhodococcus. Biodegradation 2016, 27, 277–286. [Google Scholar] [CrossRef]
- Nam, J.H.; Ventura, J.R.S.; Yeom, I.T.; Lee, Y.; Jahng, D. Structural and kinetic characteristics of 1,4-dioxane-degrading bacterial consortia containing the phylum TM7. J. Microbiol. Biotechnol. 2016, 26, 1951–1964. [Google Scholar] [CrossRef]
- Inoue, D.; Tsunoda, T.; Yamamoto, N.; Ike, M.; Sei, K. 1,4-dioxane degradation characteristics of Rhodococcus aetherivorans JCM 14343. Biodegradation 2018, 29, 301–310. [Google Scholar] [CrossRef]
- Guan, X.; Liu, F.; Wang, J.; Li, C.; Zheng, X. Mechanism of 1,4-dioxane microbial degradation revealed by 16S rRNA and metatranscriptomic analyses. Water Sci. Technol. 2018, 77, 123–133. [Google Scholar] [CrossRef]
- Yamamoto, N.; Saito, Y.; Inoue, D.; Sei, K.; Ike, M. Characterization of newly isolated Pseudonocardia sp. N23 with high 1,4-dioxane-degrading ability. J. Biosci. Bioeng. 2018, 125, 552–558. [Google Scholar] [CrossRef]
- USEPA (U.S. Environmental Protection Agency). Problem Formulation of the Risk Evaluation for 1,4-Dioxane (CASRN: 123-91-1); Office of the Chemical Safety and Pollution Prevention (OCSPP): Washington, DC, USA, 2018.
- Kim, C.G.; Seo, H.J.; Lee, B.R. Decomposition of 1,4-dioxane by advanced oxidation and biochemical process. J. Environ. Sci. Health A Toxic Hazard. Subst. Environ. Eng. 2006, 41, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, N.; Nakagawa, T.; Asano, M.; Abe, M.; Yamada, M.; Ono, Y. Ozonation combined with electrolysis of 1,4-dioxane using a two compartment electrolytic flow cell with solid electrolyte. Water Res. 2008, 42, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Li, F.; Li, M. A novel propane monooxygenase initiating degradation of 1,4-dioxane by Mycobacterium dioxanotrophicus PH-06. Environ. Sci. Technol. Lett. 2018, 5, 86–91. [Google Scholar] [CrossRef]
- USEPA (U.S. Environmental Protection Agency). Treatment Technologies for 1,4-Dioxane: Fundamentals and Field Applications; Office of Superfund Remediation and Technology Innovation (OSRTI): Cincinnati, OH, USA, 2006.
- Aoyagi, T.; Morishita, F.; Sugiyama, Y.; Ichikawa, D.; Mayumi, D.; Kikuchi, Y.; Ogata, A.; Muraoka, K.; Habe, H.; Hori, T. Identification of active and taxonomically diverse 1,4-dioxane degraders in a full-scale activated sludge system by high-sensitivity stable isotope probing. ISME J. 2018, 12, 2376–2388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, X.; Polasko, A.; Johnson, N.W.; Miao, Y.; Yang, Z.; Mahendra, S.; Gu, B. Co-contaminant effects on 1,4-dioxane biodegradation in packed soil column flow-through systems. Environ. Pollut. 2018, 243, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zenker, M.J.; Borden, R.C.; Barlaz, M.A. Mineralization of 1,4-dioxane in the presence of a structural analog. Biodegradation 2000, 11, 239–246. [Google Scholar] [CrossRef]
- Shen, W.; Chen, H.; Pan, S. Anaerobic biodegradation of 1,4-dioxane by sludge enriched with iron-reducing microorganisms. Bioresour. Technol. 2008, 99, 2483–2487. [Google Scholar] [CrossRef]
- Mahendra, S.; Alvarez-Cohen, L. Kinetics of 1,4-dioxane biodegradation by monooxygenase-expressing bacteria. Environ. Sci. Technol. 2006, 40, 5435–5442. [Google Scholar] [CrossRef]
- Li, M.; Liu, Y.; He, Y.; Mathieu, J.; Hatton, J.; DiGuiseppi, W.; Alvarez, P.J.J. Hindrance of 1,4-dioxane biodegradation in microcosms biostimulated with inducing or non-inducing auxiliary substrates. Water Res. 2017, 112, 217–225. [Google Scholar] [CrossRef]
- Mahendra, S.; Grostern, A.; Alvarez-Cohen, L. The impact of chlorinated solvent co-contaminants on the biodegradation kinetics of 1,4-dioxane. Chemosphere 2013, 91, 88–92. [Google Scholar] [CrossRef]
- Pornwongthong, P.; Mulchandani, A.; Gedalanga, P.B.; Mahendra, S. Transition metals and organic ligands influence biodegradation of 1,4-dioxane. Appl. Biochem. Biotechnol. 2014, 173, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gedalanga, P.B.; Mahendra, S. Biodegradation kinetics of 1,4-dioxane in chlorinated solvent mixtures. Environ. Sci. Technol. 2016, 50, 9599–9607. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, H.; Shen, D. Multi-substrate biodegradation interaction of 1,4-dioxane and BTEX mixtures by Acinetobacter baumannii DD1. Biodegradation 2016, 27, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Barajas-Rodriquez, F.J.; Freedman, D.L. Aerobic biodegradation kinetics for 1,4-dioxane under metabolic and cometabolic conditions. J. Hazard. Mater. 2018, 350, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Mason, O.U.; Lowe, A.; Zhou, C.; Chen, G.; Tang, Y. Microbial community analysis provides insights into the effects of tetrahydrofuran on 1,4-dioxane biodegradation. Appl. Environ. Microbiol. 2019, 85, e00244. [Google Scholar] [CrossRef]
- Monferrán, M.V.; Echenique, J.R.; Wunderlin, D.A. Degradation of chlorobenzenes by a strain of Acidovorax avenae isolated from a polluted aquifer. Chemosphere 2005, 61, 98–106. [Google Scholar] [CrossRef]
- Parales, R.E.; Adamus, J.E.; White, N.; May, H.D. Degradation of 1,4-dioxane by an Actinomycete in pure culture. Appl. Environ. Microbiol. 1994, 60, 4527–4530. [Google Scholar]
- Mahendra, S.; Alvarez-Cohen, L. Pseudonocardia dioxanivorans sp. nov., a novel Actinomycete that grows on 1,4-dioxane. Int. J. Syst. Evol. Microbiol. 2005, 55, 593–598. [Google Scholar] [CrossRef]
- Satola, B.; Wübbeler, J.H.; Steinbüchel, A. Metabolic characteristics of the species Variovorax paradoxus. Appl. Microbiol. Biotechnol. 2013, 97, 541–560. [Google Scholar] [CrossRef]
- Sun, B.; Ko, K.; Ramsay, J.A. Biodegradation of 1,4-dioxane by a Flavobacterium. Biodegradation 2011, 22, 651–659. [Google Scholar] [CrossRef]
- Arulazhagan, P.; Yeom, I.T.; Sivaraman, C.; Srikanth, M.; Rajesh, B.J. Role of nutrients on biodegradation of 1,4-dioxane by a bacterial consortium enriched from industrial sludge. Adv. Environ. Biol. 2013, 7, 2081–2090. [Google Scholar]
- He, Y.; Mathieu, J.; da Silva, M.L.B.; Li, M.; Alvarez, P.J.J. 1,4-dioxane-degrdaing consortia can be enriched from uncontaminated soils: Prevalence of Mycobacterium and soluble di-iron monooxygenase genes. Microb. Biotechnol. 2018, 11, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Chien, M.-F.; Suto, K.; Inoue, C. Analysis of stable 1,2-dichlorobenzene-degradaing enrichments and two newly isolated degrading strains, Acidovoax sp. sk40 and Ralstonia sp. sk41. Appl. Microbiol. Biotechnol. 2017, 101, 6821–6828. [Google Scholar] [CrossRef] [PubMed]
- Grostern, A.; Sales, C.M.; Zhang, W.Q.; Erbilgin, O.; Alvarez-Cohen, L. Glyoxylate metabolism is a key feature of the metabolic degradation of 1,4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Appl. Environ. Microbiol. 2012, 78, 3298–3308. [Google Scholar] [CrossRef] [PubMed]
- Coleman, N.V.; Bui, N.B.; Holmes, A.J. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ. Microbiol. 2006, 8, 1228–1239. [Google Scholar] [CrossRef]
- Han, J.-I.; Choi, H.-K.; Lee, S.-W.; Orwin, P.M.; Kim, J.; LaRoe, S.L.; Kim, T.-G.; O’Neil, J.; Leadbetter, J.R.; Lee, S.Y.; et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011, 193, 1183–1190. [Google Scholar] [CrossRef]
- Snellinx, Z.; Taghavi, S.; Vangronsveld, J.; Lelie, D. Microbial consortia that degrade 2,4-DNT by interspecies metabolism: Isolation and characterization. Biodegradation 2003, 14, 19–29. [Google Scholar] [CrossRef]
- Zaitsev, G.M.; Uotila, J.S.; Häggblom, M.M. Biodegradation of methyl tert-butyl ether by cold-adapted mixed and pure bacterial cultures. Appl. Microbiol. Biotechnol. 2007, 74, 1092–1102. [Google Scholar] [CrossRef]
- Da Silva, M.L.B.; Woroszylo, C.; Castillo, N.F.; Adamson, D.T.; Alvarez, P.J.J. Associating potential 1,4-dioxane biodegradation activity with groundwater geochemical parameters at four different contaminated sites. J. Environ. Manag. 2018, 206, 60–64. [Google Scholar] [CrossRef]
- Ziesack, M.; Gibson, T.; Oliver, J.K.W.; Shumaker, A.M.; Hsu, B.B.; Riglar, D.T.; Giessen, T.W.; DiBenedetto, N.V.; Bry, L.; Way, J.C.; et al. Engineered interspecies amino acid cross-feeding increases population evenness in a synthetic bacterial consortium. mSystems 2019, 4, e00352. [Google Scholar] [CrossRef]

| Purpose | Target Gene | Primer Name | Primer Sequence (5ʹ → 3ʹ) |
|---|---|---|---|
| Isolation | 16S rRNA | 27F | AGAGTTTGATCCTGGCTCAG |
| 1492R | TACGGYTACCTTGTTACGACTT | ||
| RISA | 16S-23S rRNA | ITSF ITSR | GTCGTAACAAGGTAGCCGTA GCCAAGGCATCCACC |
| NGS (1st PCR) | 16S rRNA (V4) | 515F_MIX 806R_MIX | ACACTCTTTCCCTACACGACGCTCTTCCGATCT-NNNNN-GTGCCAGCMGCCGCGGTAA GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-NNNNN-GGACTACHVGGGTWTCTAAT |
| Isolated Strains | Family (Class) | Closest Relative (Accession No.) | Similarity with the Closest Relative (%) | Accession No. |
|---|---|---|---|---|
| Pseudonocardia sp. TS06 | Pseudonocardiaceae (Actinobacteria) | Pseudonocardia dioxanivorans CB1190 (NR_074465) | 99.79 | MN715853 |
| Variovorax sp. TS13 | Comamonadaceae (β-proteobacteria) | Variovorax paradoxus NBRC 15149 (NR_113736) | 99.50 | MN715854 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tusher, T.R.; Shimizu, T.; Inoue, C.; Chien, M.-F. Enrichment and Analysis of Stable 1,4-dioxane-Degrading Microbial Consortia Consisting of Novel Dioxane-Degraders. Microorganisms 2020, 8, 50. https://doi.org/10.3390/microorganisms8010050
Tusher TR, Shimizu T, Inoue C, Chien M-F. Enrichment and Analysis of Stable 1,4-dioxane-Degrading Microbial Consortia Consisting of Novel Dioxane-Degraders. Microorganisms. 2020; 8(1):50. https://doi.org/10.3390/microorganisms8010050
Chicago/Turabian StyleTusher, Tanmoy Roy, Takuya Shimizu, Chihiro Inoue, and Mei-Fang Chien. 2020. "Enrichment and Analysis of Stable 1,4-dioxane-Degrading Microbial Consortia Consisting of Novel Dioxane-Degraders" Microorganisms 8, no. 1: 50. https://doi.org/10.3390/microorganisms8010050
APA StyleTusher, T. R., Shimizu, T., Inoue, C., & Chien, M.-F. (2020). Enrichment and Analysis of Stable 1,4-dioxane-Degrading Microbial Consortia Consisting of Novel Dioxane-Degraders. Microorganisms, 8(1), 50. https://doi.org/10.3390/microorganisms8010050