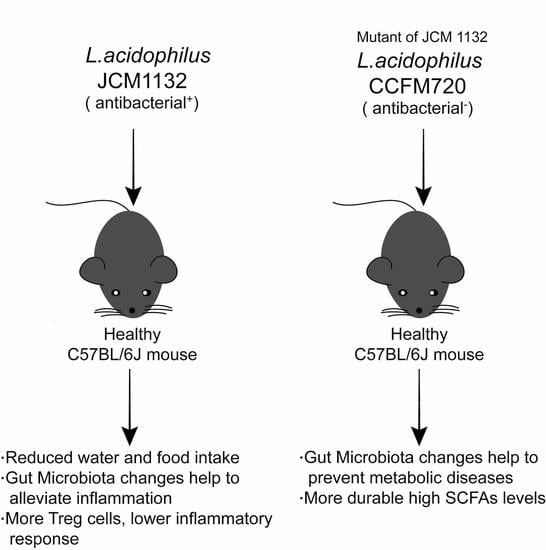
Lactobacillus acidophilus JCM 1132 Strain and Its Mutant with Different Bacteriocin-Producing Behaviour Have Various In Situ Effects on the Gut Microbiota of Healthy Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Measurement of Antibacterial Activities and Spectra
2.2. Sensitivity of Antibacterial Substances in CFS to Catalase, Protease and Heat
2.3. Biological Characteristics of the Two Tested Strains
2.3.1. Growth Curve, Generation Time and Adhesion Assay
2.3.2. Determination of the Tolerance of Two Strains to Simulated GI Tract Conditions
2.4. Genome Sequencing and Assemblies
2.5. MALDI-TOF MS
2.6. Animals and Experimental Design
2.7. Histomorphological Analysis
2.8. Biochemical Measurements
2.9. Flow Cytometry
2.10. 16S rDNA Amplicon Sequencing
2.11. Determination of SCFAs in Mice Faeces
2.12. Statistical Analysis
3. Results
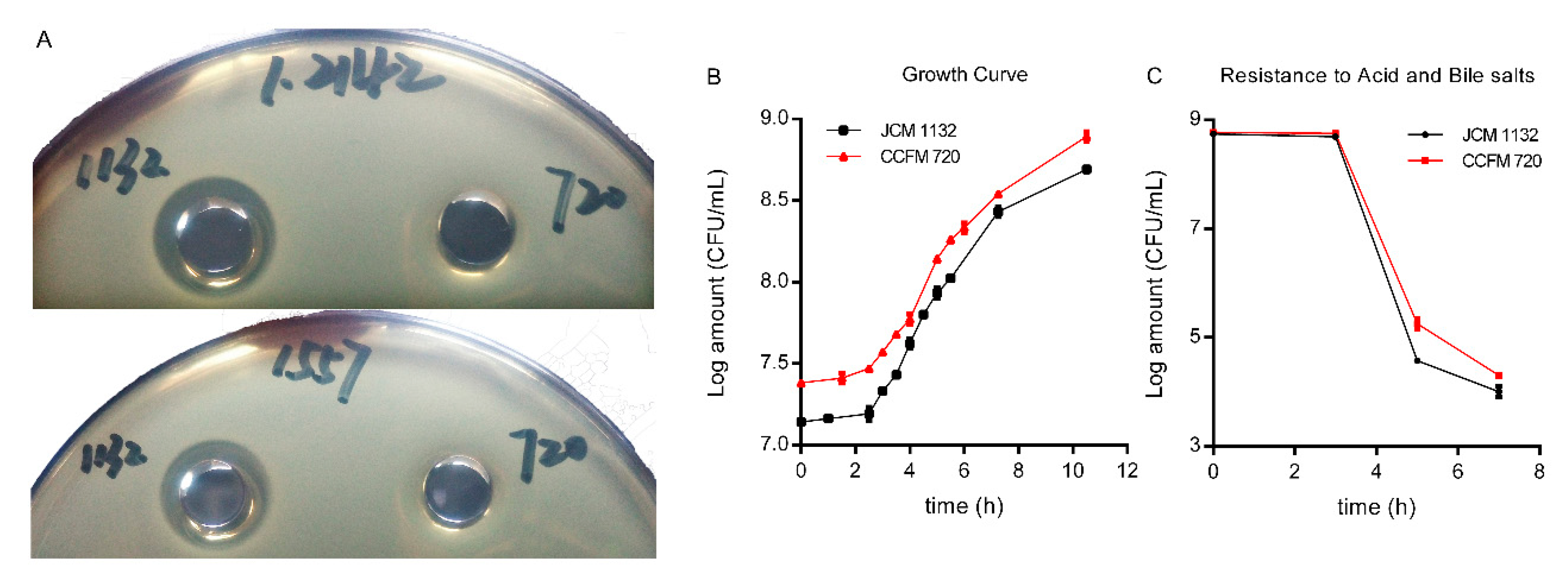
3.1. Loss of CCFM 720 Antimicrobial Activity against L. delbrueckii subsp. lactis
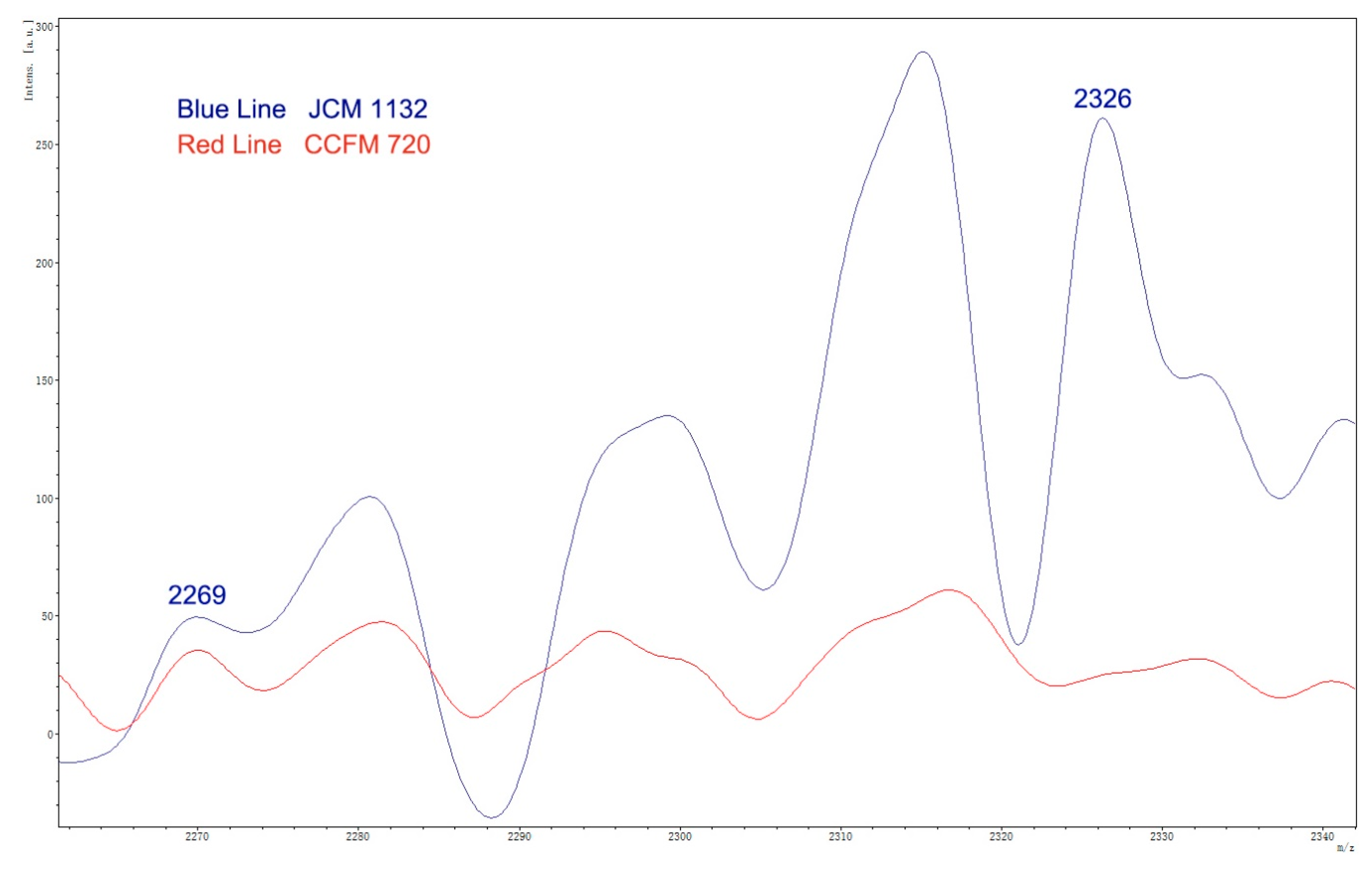
3.2. Absence of Antimicrobial Activity in CCFM720 Was Due to the Loss of Bacteriocin Activity
3.3. Genome Analysis
3.4. JCM 1132 and CCFM 720 Exhibited Similar Growth, Adhesion Characteristics and Resistance to Gastric acid and Bile Salts
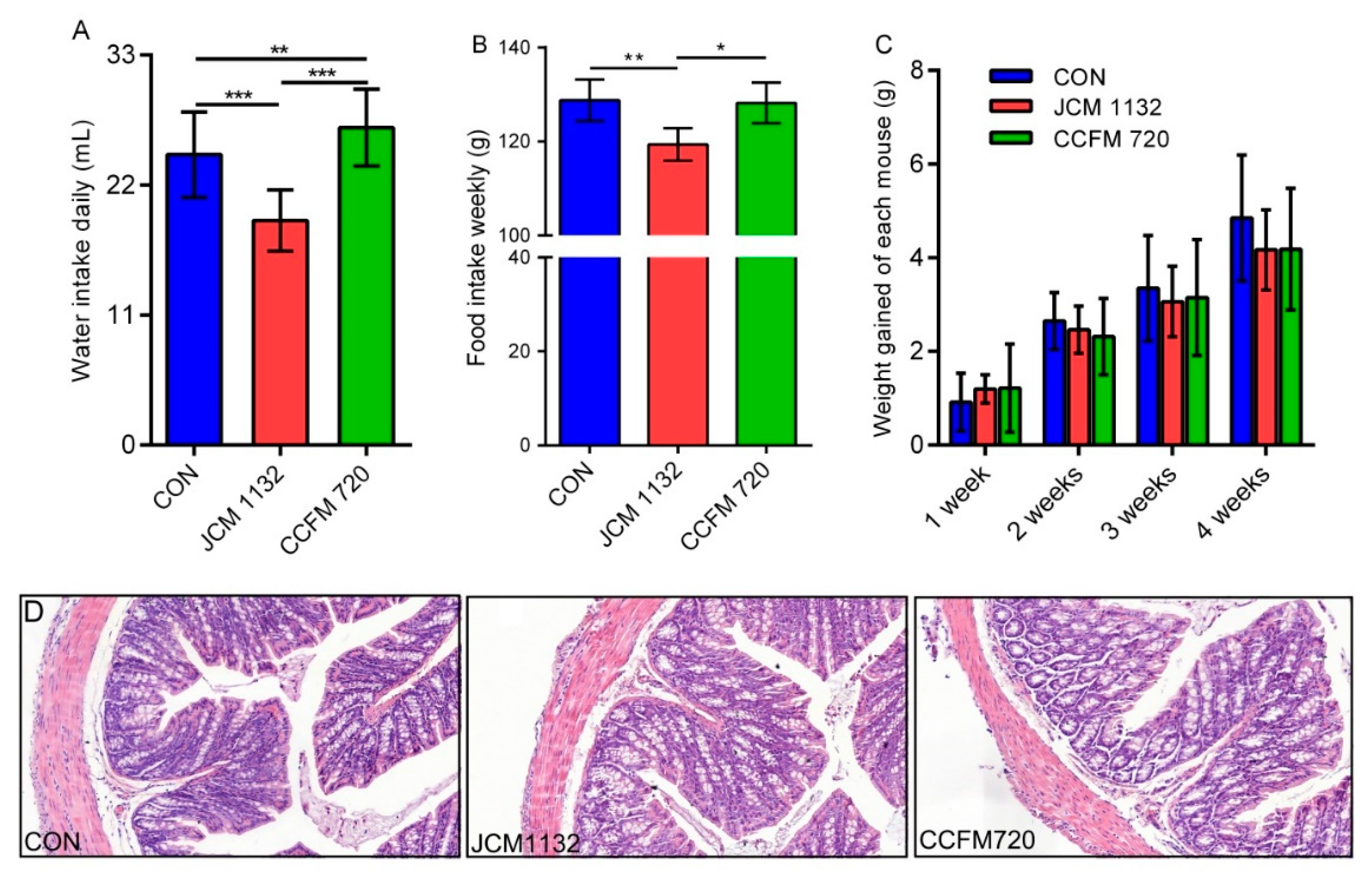
3.5. JCM 1132 and CCFM 720 Had Different Physiological Effects on Healthy Mice
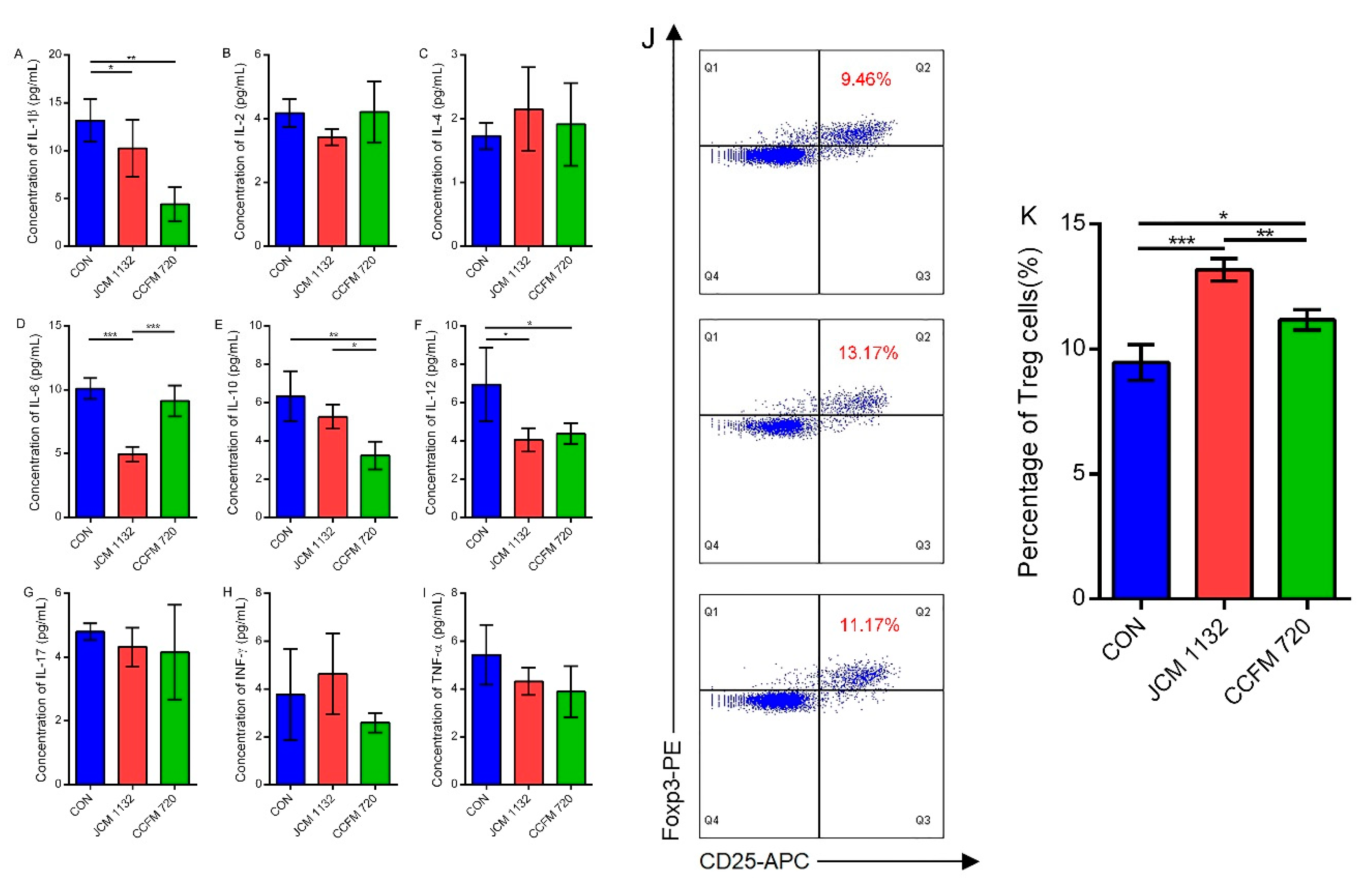
3.6. JCM1132 More Effectively Promoted an Immunosuppressive Response in Healthy Mice
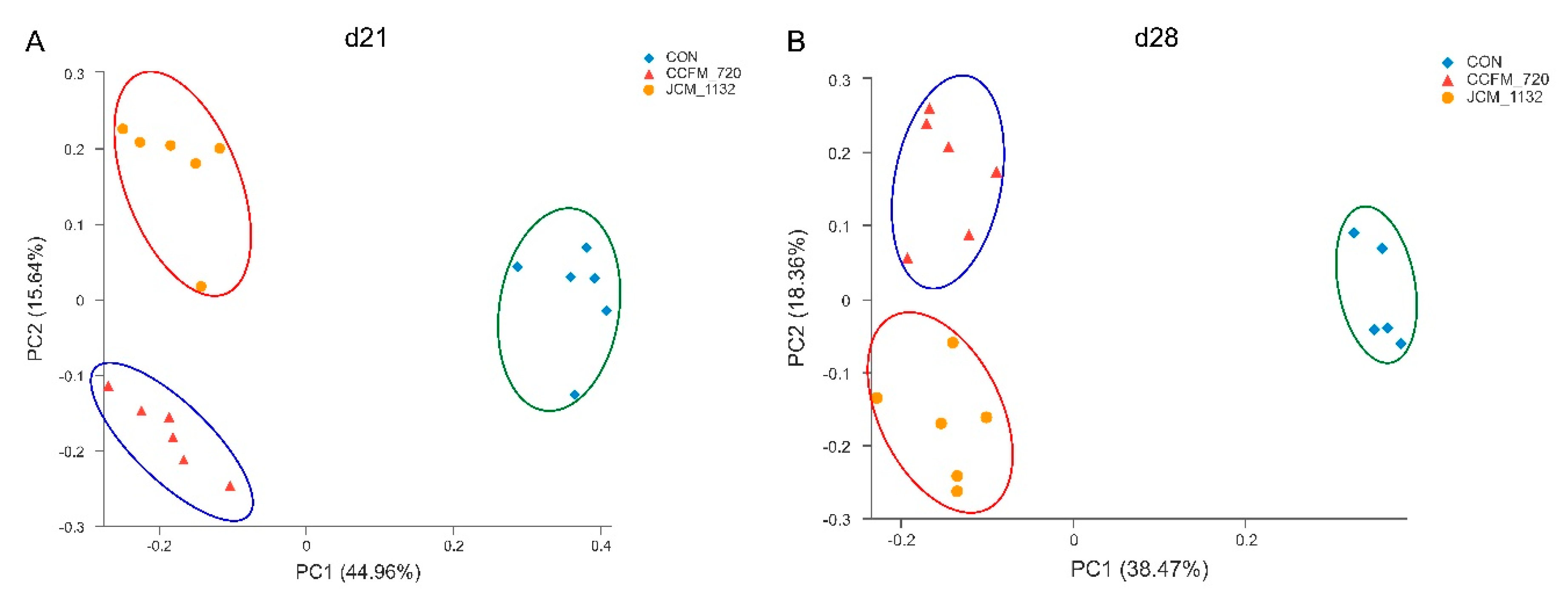
3.7. JCM 1132 and CCFM 720 Had Different Effects on the β Diversity of Faecal Microbiota
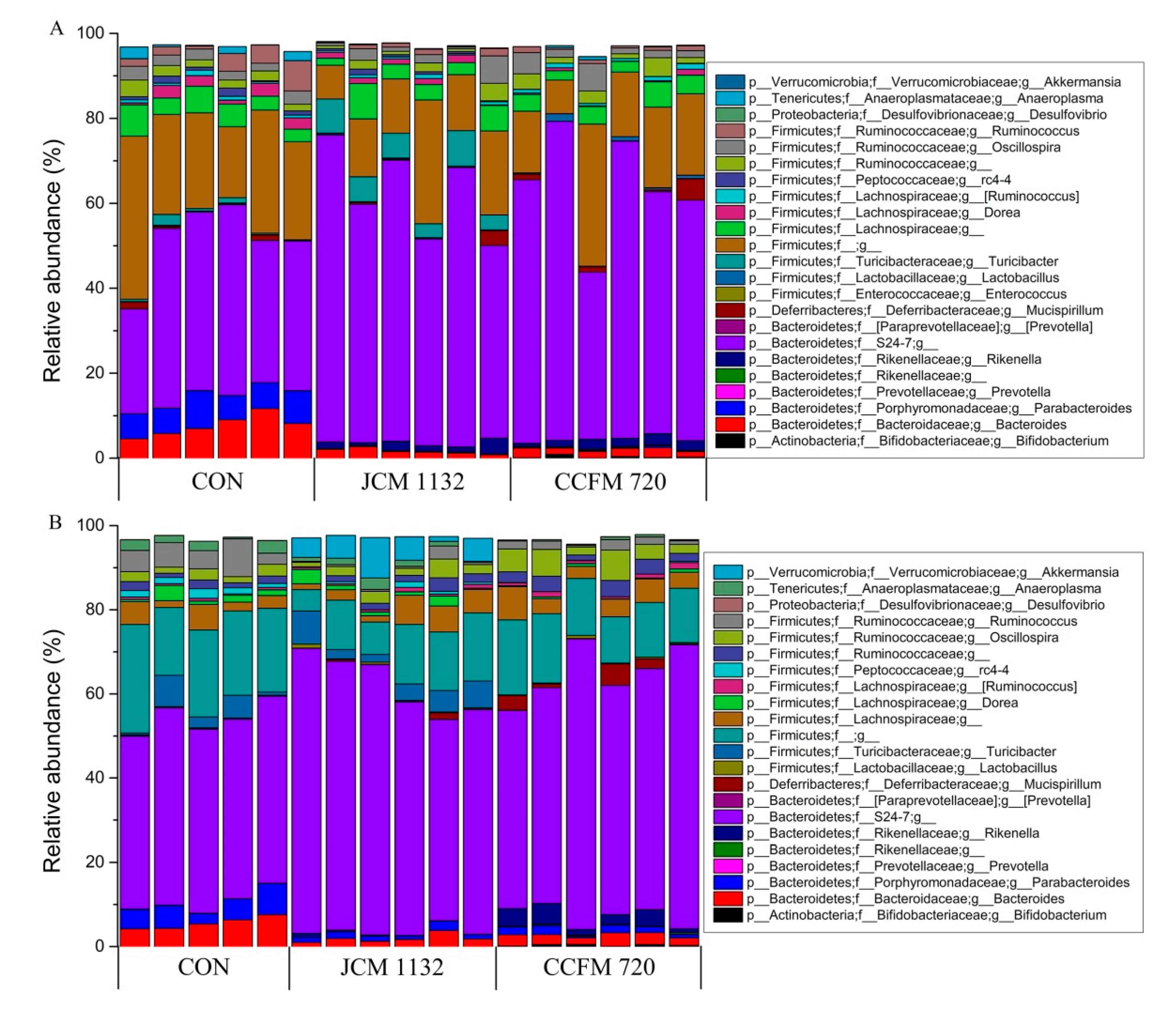
3.8. Treatment with JCM 1132 and CCFM 720 Affected Gut Microbiota Patterns at the Phylum Level
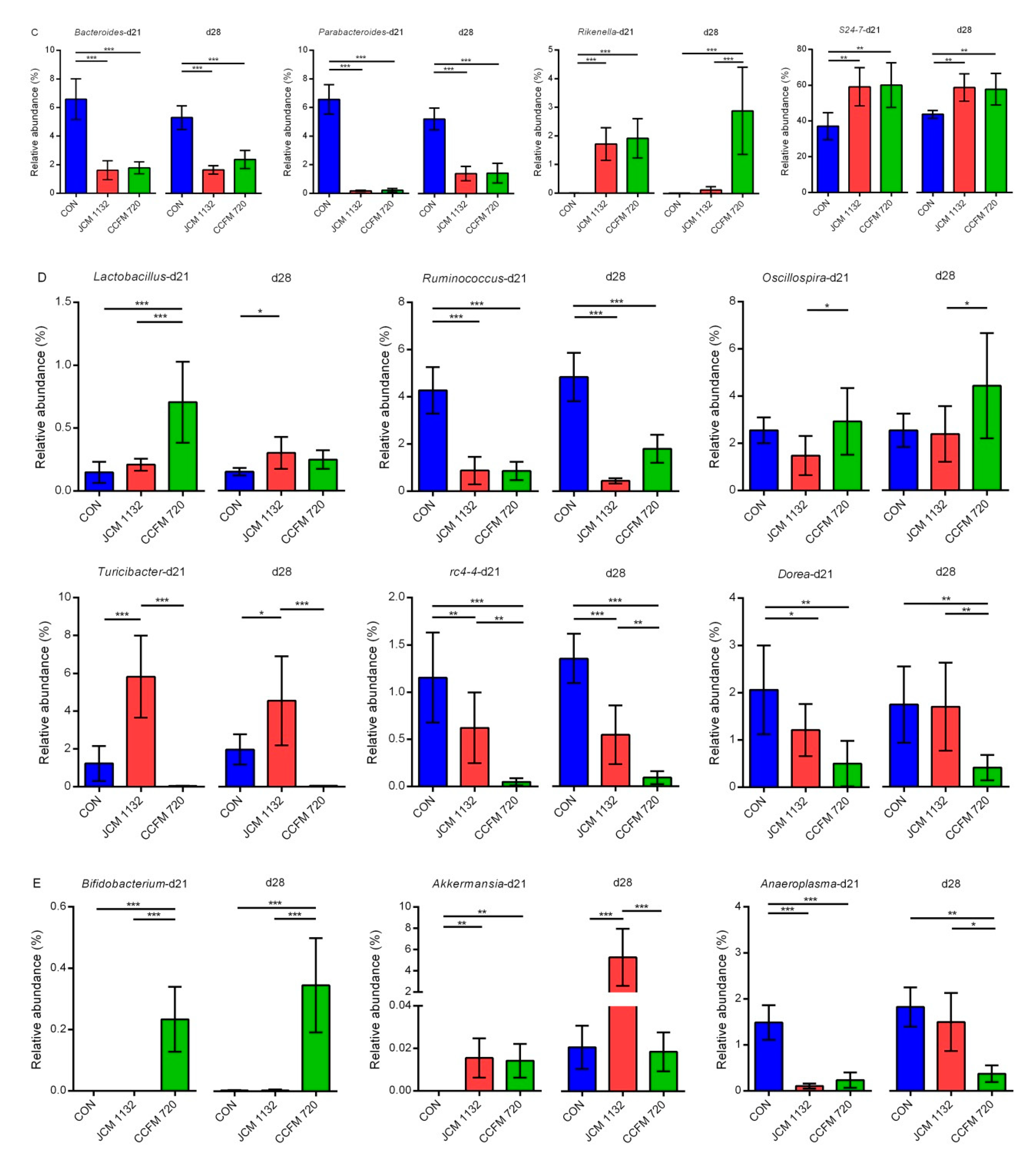
3.9. Treatment with JCM 1132 and CCFM 720 Affected the Gut Microbiota Patterns at the Genus Level
3.9.1. Relative Abundance of Selected Genera in Bacteroidetes
3.9.2. Relative Abundance of Selected Genus in Firmicutes
3.9.3. Relative Abundance of Other Genera
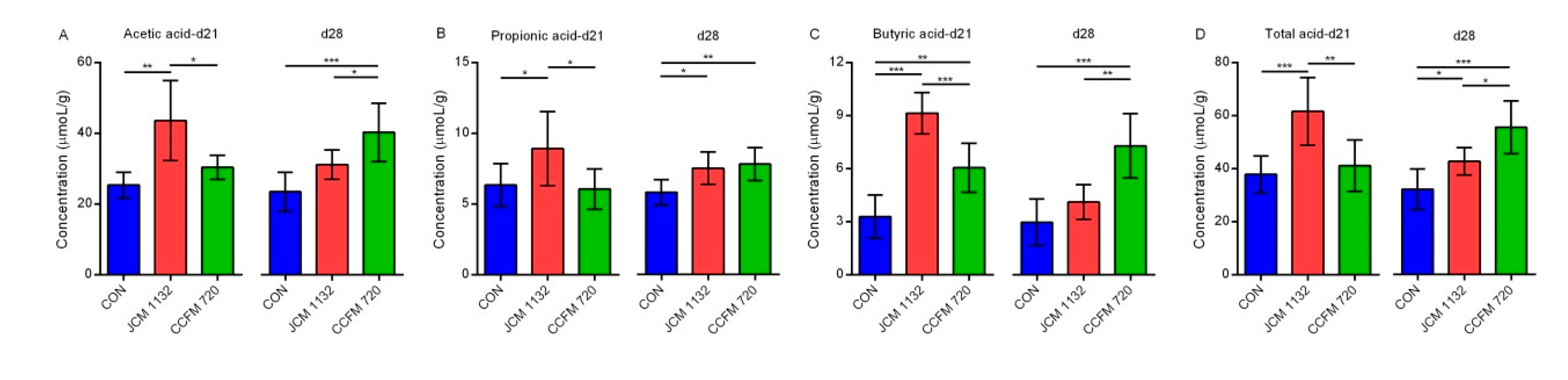
3.10. Administration of JCM 1132 and CCFM 720 Influenced the Levels and Patterns of SCFAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- DuPont, A.W.; DuPont, H.L. The intestinal microbiota and chronic disorders of the gut. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.S. Mucosal immunity and the microbiome. Ann. Am. Thorac. Soc. 2014, 11, S28–S32. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Bayer, E.A.; Rincon, M.T.; Lamed, R.; White, B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; Mayer, M.J.; Cotter, P.D.; Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 2019, 10, 1–21. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; De Giori, G.S.; Sesma, F.; Van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Maynard, C.L.; Elson, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef]
- Schluter, J.; Foster, K.R. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012, 10, e1001424. [Google Scholar] [CrossRef]
- Zaibi, M.S.; Stocker, C.J.; O′Dowd, J.; Davies, A.; Bellahcene, M.; Cawthorne, M.A.; Brown, A.J.; Smith, D.M.; Arch, J.R. Roles of gpr41 and gpr43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett. 2010, 584, 2381–2386. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate gpr41 and gpr43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406.e310. [Google Scholar] [CrossRef] [PubMed]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar] [CrossRef] [PubMed]
- Kailasapathy, K.; Chin, J. Survival and therapeutic potential of probiotic organisms with reference to lactobacillus acidophilus and bifidobacterium spp. Immunol. Cell Biol. 2000, 78, 80–88. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, E.F.; Gardiner, G.E.; O’Connor, P.M.; Mills, S.; Ross, R.P.; Hill, C. Characterization of enterocin-and salivaricin-producing lactic acid bacteria from the mammalian gastrointestinal tract. FEMS Microbiol. Lett. 2009, 291, 24–34. [Google Scholar] [CrossRef]
- Lakshminarayanan, B.; Guinane, C.; O’Connor, P.; Coakley, M.; Hill, C.; Stanton, C.; O’Toole, P.; Ross, R. Isolation and characterization of bacteriocin-producing bacteria from the intestinal microbiota of elderly irish subjects. J. Appl. Microbiol. 2013, 114, 886–898. [Google Scholar] [CrossRef]
- Jena, P.K.; Trivedi, D.; Chaudhary, H.; Sahoo, T.K.; Seshadri, S. Bacteriocin pj4 active against enteric pathogen produced by lactobacillus helveticus pj4 isolated from gut microflora of wistar rat (rattus norvegicus): Partial purification and characterization of bacteriocin. Appl. Biochem. Biotechnol. 2013, 169, 2088–2100. [Google Scholar] [CrossRef]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Riboulet-Bisson, E.; Sturme, M.H.; Jeffery, I.B.; O’Donnell, M.M.; Neville, B.A.; Forde, B.M.; Claesson, M.J.; Harris, H.; Gardiner, G.E.; Casey, P.G. Effect of lactobacillus salivarius bacteriocin abp118 on the mouse and pig intestinal microbiota. PLoS ONE 2012, 7, e31113. [Google Scholar] [CrossRef]
- Kwok, L.; Guo, Z.; Zhang, J.; Wang, L.; Qiao, J.; Hou, Q.; Zheng, Y.; Zhang, H. The impact of oral consumption of lactobacillus plantarum p-8 on faecal bacteria revealed by pyrosequencing. Benef. Microbes 2015, 6, 405–413. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Yi, H.; Han, X.; Gao, W.; Chi, C.; Song, W.; Li, H.; Liu, C. A novel enterocin t1 with anti-pseudomonas activity produced by enterococcus faecium t1 from chinese tibet cheese. World J. Microbiol. Biotechnol. 2016, 32, 21. [Google Scholar] [CrossRef]
- Todorov, S.D.; Furtado, D.N.; Saad, S.M.I.; Tome, E.; Franco, B.D.G.d.M. Potential beneficial properties of bacteriocin-producing lactic acid bacteria isolated from smoked salmon. J. Appl. Microbiol. 2011, 110, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Ortolani, M.B.T.; Yamazi, A.K.; Moraes, P.M.; Viçosa, G.N.; Nero, L.A. Microbiological quality and safety of raw milk and soft cheese and detection of autochthonous lactic acid bacteria with antagonistic activity against listeria monocytogenes, salmonella spp., and staphylococcus aureus. Foodborne Pathog. Dis. 2010, 7, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.; Dicks, L. Screening for bacteriocin-producing lactic acid bacteria from boza, a traditional cereal beverage from bulgaria: Comparison of the bacteriocins. Process Biochem. 2006, 41, 11–19. [Google Scholar] [CrossRef]
- Zhai, Q.; Yin, R.; Yu, L.; Wang, G.; Tian, F.; Yu, R.; Zhao, J.; Liu, X.; Chen, Y.Q.; Zhang, H. Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control 2015, 54, 23–30. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. Bagel4: A user-friendly web server to thoroughly mine ripps and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. Patric, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef]
- O’Connor, P.M.; O’Shea, E.F.; Guinane, C.M.; O’Sullivan, O.; Cotter, P.D.; Ross, R.P.; Hill, C. Nisin h is a new nisin variant produced by the gut-derived strain streptococcus hyointestinalis dpc6484. Appl. Environ. Microbiol. 2015, 81, 3953–3960. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Xu, J.; Hong, J.; Le, Q. Pretreatment of rapamycin before allogenic corneal transplant promotes graft survival through increasing cd4 (+) cd25 (+) foxp3 (+) regulatory t cells. Exp. Clin. Transplant. Off. J. Middle East Soc. Organ Transplant. 2013, 11, 56–62. [Google Scholar] [CrossRef]
- Wang, G.; He, Y.; Jin, X.; Zhou, Y.; Chen, X.; Zhao, J.; Zhang, H.; Chen, W. The effect of co-infection of food-borne pathogenic bacteria on the progression of campylobacter jejuni infection in mice. Front. Microbiol. 2018, 9, 1977. [Google Scholar] [CrossRef]
- Tahara, T.; Oshimura, M.; Umezawa, C.; Kanatani, K. Isolation, partial characterization, and mode of action of acidocin j1132, a two-component bacteriocin produced by lactobacillus acidophilus jcm 1132. Appl. Environ. Microbiol. 1996, 62, 892–897. [Google Scholar]
- Stephani, J.; Radulovic, K.; Niess, J.H. Gut microbiota, probiotics and inflammatory bowel disease. Arch. Immunol. Ther. Exp. 2011, 59, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Li, S.; Gan, R.Y.; Zhou, T.; Xu, D.P.; Li, H.B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; He, J.; Gao, N.; Lu, X.; Li, M.; Wu, X.; Liu, Z.; Jin, Y.; Liu, J.; Xu, J. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci. Rep. 2017, 7, 45176. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ibarreche, M.; Castellano, P.; Leclercq, A.; Vignolo, G. Control of listeria monocytogenes biofilms on industrial surfaces by the bacteriocin-producing lactobacillus sakei crl1862. FEMS Microbiol. Lett. 2016, 363, fnw118. [Google Scholar] [CrossRef] [PubMed]
- Barrena-Gonzalez, C.; Huot, E.; Petitdemange, H. Mode of action of a bacteriocin (j46) produced by lactococcus lactis subsp. Cremoris j46. J. Food Prot. 1996, 59, 955–962. [Google Scholar] [CrossRef]
- Gao, P.; Ma, C.; Sun, Z.; Wang, L.; Huang, S.; Su, X.; Xu, J.; Zhang, H. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 2017, 5, 91. [Google Scholar] [CrossRef]
- Tomita, H.; Fujimoto, S.; Tanimoto, K.; Ike, Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the enterococcus faecalis pheromone-responsive conjugative plasmid ppd1. J. Bacteriol. 1997, 179, 7843–7855. [Google Scholar] [CrossRef]
- Chang, Y.H.; Kim, J.K.; Kim, H.J.; Kim, W.Y.; Kim, Y.B.; Park, Y.H. Selection of a potential probiotic lactobacillus strain and subsequent in vivo studies. Antonie Van Leeuwenhoek 2001, 80, 193–199. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Bernbom, N.; Jelle, B.; Brogren, C.-H.; Vogensen, F.K.; Nørrung, B.; Licht, T.R. Pediocin pa-1 and a pediocin producing lactobacillus plantarum strain do not change the hma rat microbiota. Int. J. Food Microbiol. 2009, 130, 251–257. [Google Scholar] [CrossRef]
- Kasuga, G.; Tanaka, M.; Harada, Y.; Nagashima, H.; Yamato, T.; Wakimoto, A.; Arakawa, K.; Kawai, Y.; Kok, J.; Masuda, T. Homologous expression and characterization of gassericin t and gassericin s, a novel class iib bacteriocin produced by lactobacillus gasseri la327. Appl. Environ. Microbiol. 2019, 85, e02815–e02818. [Google Scholar] [CrossRef] [PubMed]
- Umu, Ö.C.; Bäuerl, C.; Oostindjer, M.; Pope, P.B.; Hernández, P.E.; Pérez-Martínez, G.; Diep, D.B. The potential of class ii bacteriocins to modify gut microbiota to improve host health. PLoS ONE 2016, 11, e0164036. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Bercik, P.; Verma-Gandhu, M.; Huang, X.-X.; Blennerhassett, P.; Jackson, W.; Mao, Y.; Wang, L.; Rochat, F.; Collins, S.M. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 2006, 55, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut microbiome: Profound implications for diet and disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Fujio-Vejar, S.; Vasquez, Y.; Morales, P.; Magne, F.; Vera-Wolf, P.; Ugalde, J.A.; Navarrete, P.; Gotteland, M. The gut microbiota of healthy chilean subjects reveals a high abundance of the phylum verrucomicrobia. Front. Microbiol. 2017, 8, 1221. [Google Scholar] [CrossRef]
- Collado, M.C.; Derrien, M.; Isolauri, E.; de Vos, W.M.; Salminen, S. Intestinal integrity and akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770. [Google Scholar] [CrossRef]
- Derrien, M.; Collado, M.C.; Ben-Amor, K.; Salminen, S.; de Vos, W.M. The mucin degrader akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008, 74, 1646–1648. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the akkermansia spp. Population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ma, F.; Wang, G.; Wang, Y.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacteria attenuate the development of metabolic disorders, with inter-and intra-species differences. Food Funct. 2018, 9, 3509–3522. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Feng, B.; Li, P.; Tang, Z.; Wang, L. Microflora disturbance during progression of glucose intolerance and effect of sitagliptin: An animal study. J. Diabetes Res. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, V.; Kaakoush, N.O.; Maloney, C.A.; Raipuria, M.; Huinao, K.D.; Mitchell, H.M.; Morris, M.J. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS ONE 2015, 10, e0126931. [Google Scholar] [CrossRef] [PubMed]
- Millette, M.; Cornut, G.; Dupont, C.; Shareck, F.; Archambault, D.; Lacroix, M. Capacity of human nisin-and pediocin-producing lactic acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl. Environ. Microbiol. 2008, 74, 1997–2003. [Google Scholar] [CrossRef]
- Liu, W.; Ren, P.; He, S.; Xu, L.; Yang, Y.; Gu, Z.; Zhou, Z. Comparison of adhesive gut bacteria composition, immunity, and disease resistance in juvenile hybrid tilapia fed two different lactobacillus strains. Fish Shellfish Immunol. 2013, 35, 54–62. [Google Scholar] [CrossRef]
- Oh, P.L.; Benson, A.K.; Peterson, D.A.; Patil, P.B.; Moriyama, E.N.; Roos, S.; Walter, J. Diversification of the gut symbiont lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010, 4, 377–387. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Y.; Wang, Y.; Li, Y.; Zheng, H.; Ma, F.; Ma, C.; Zhang, X.; Lu, B.; Xie, Z. The effect of probiotics and polysaccharides on the gut microbiota composition and function of weaned rats. Food Funct. 2018, 9, 1864–1877. [Google Scholar] [CrossRef]
- Holzapfel, W.; Geisen, R.; Schillinger, U. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int. J. Food Microbiol. 1995, 24, 343–362. [Google Scholar] [CrossRef]
- Magnusson, J.; Schnürer, J. Lactobacillus coryniformis subsp. Coryniformis strain si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001, 67, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Oki, K.; Toyama, M.; Banno, T.; Chonan, O.; Benno, Y.; Watanabe, K. Comprehensive analysis of the fecal microbiota of healthy japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol. 2016, 16, 284. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, H.; Shi, Q.; Wang, N.; Zhang, Z.; Xiong, C.; Liu, J.; Chen, Y.; Jiang, L.; Jiang, Q. Effects of oral florfenicol and azithromycin on gut microbiota and adipogenesis in mice. PLoS ONE 2017, 12, e0181690. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Chang, W.T.; Li, S.; Wu, J.C.; Badmeav, V.; Ho, C.T.; Pan, M.H. Citrus peel extracts attenuated obesity and modulated gut microbiota in mice with high-fat diet-induced obesity. Food Funct. 2018, 9, 3363–3373. [Google Scholar] [CrossRef]
- Sun, H.; Wang, N.; Cang, Z.; Zhu, C.; Zhao, L.; Nie, X.; Cheng, J.; Xia, F.; Zhai, H.; Lu, Y. Modulation of microbiota-gut-brain axis by berberine resulting in improved metabolic status in high-fat diet-fed rats. Obes. Facts 2016, 9, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Brahe, L.K.; Le Chatelier, E.; Prifti, E.; Pons, N.; Kennedy, S.; Hansen, T.; Pedersen, O.; Astrup, A.; Ehrlich, S.; Larsen, L.H. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes 2015, 5, e159. [Google Scholar] [CrossRef]
- Rajilić–Stojanović, M.; Biagi, E.; Heilig, H.G.; Kajander, K.; Kekkonen, R.A.; Tims, S.; de Vos, W.M. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1792–1801. [Google Scholar] [CrossRef]
- Shen, X.J.; Rawls, J.F.; Randall, T.A.; Burcall, L.; Mpande, C.; Jenkins, N.; Jovov, B.; Abdo, Z.; Sandler, R.S.; Keku, T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010, 1, 138–147. [Google Scholar] [CrossRef]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R. A novel ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Tun, H.M.; Konya, T.; Takaro, T.K.; Brook, J.R.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E. Exposure to household furry pets influences the gut microbiota of infants at 3–4 months following various birth scenarios. Microbiome 2017, 5, 40. [Google Scholar] [CrossRef]
- Granado-Serrano, A.B.; Martín-Garí, M.; Sánchez, V.; Solans, M.R.; Berdún, R.; Ludwig, I.A.; Rubió, L.; Vilaprinyó, E.; Portero-Otín, M.; Serrano, J. Faecal bacterial and short-chain fatty acids signature in hypercholesterolemia. Sci. Rep. 2019, 9, 1772. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Ross, R.P.; O’Toole, P.W.; Shanahan, F.; Cotter, P.D. Targeting the microbiota to address diet-induced obesity: A time dependent challenge. PLoS ONE 2013, 8, e65790. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Tian, W.J.; Kwok, L.-Y.; Wang, Y.L.; Shang, Y.N.; Menghe, B.; Wang, J.G. Effects of microencapsulated lactobacillus plantarum lip-1 on the gut microbiota of hyperlipidaemic rats. Br. J. Nutr. 2017, 118, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Baker, S.S.; Nugent, C.A.; Tsompana, M.; Cai, L.; Wang, Y.; Buck, M.J.; Genco, R.J.; Baker, R.D.; Zhu, R. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol. Genom. 2018, 50, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Shang, L.; Zeng, X.; Li, N.; Liu, H.; Cai, S.; Huang, S.; Wang, G.; Wang, Y.; Song, Q. Risks related to high-dosage recombinant antimicrobial peptide microcin j25 in mice model: Intestinal microbiota, intestinal barrier function, and immune regulation. J. Agric. Food Chem. 2018, 66, 11301–11310. [Google Scholar] [CrossRef]



| Species | Strain | Source a | Culture Conditions |
|---|---|---|---|
| Tested strains | |||
| Lactobacillus acidophilus | JCM 1132 | JCM | 37 °C, MRS, Anaerobic |
| CCFM 720 | CCFM | 37 °C, MRS, Anaerobic | |
| Indicator strains | |||
| Lactobacillus plantarum | S8 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus coryniformis | S16 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus plantarum | S17 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus fermentum | S40 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus casei | W3 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus plantarum | W6 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus plantarum | W11 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus plantarum | W13 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus plantarum | W19 | CCFM | 37 °C, MRS, Aerobic |
| Entero-invasive Escherichia coli | ATCC 43893 | CCFM | 37 °C, TSB, Aerobic |
| Staphylococcus aureus | S. aureus | CCFM | 37 °C, TSB, Aerobic |
| Salmonella typhimurium | SL1344 | CCFM | 37 °C, TSB, Aerobic |
| Enterococcus faecalis | E. faecalis | CCFM | 37 °C, TSB, Aerobic |
| Lactobacillus reuteri | 9-5 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus reuteri | L103 | CCFM | 37 °C, MRS, Aerobic |
| Lactococcus Lactis | N5 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus rhamnosus | LGG | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus salivarius | ZX5 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus fermentum | D2 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus acidophilus | FFJND6-L5 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus crispatus | FHUBES1M18 | CCFM | 37 °C, MRS, Aerobic |
| Lactobacillus crispatus | FHUBES1M24 | CCFM | 37 °C, MRS, Aerobic |
| Listeria monocytogenes | ATCC 19114 | ATCC | 37 °C, TSB, Aerobic |
| Streptococcus mutans | ATCC 25175 | ATCC | 37 °C, TSB, Aerobic |
| Lactobacillus delbrueckii subsp. lactis | JCM 1557 | JCM | 37 °C, MRS, Aerobic |
| Lactobacillus delbrueckii subsp. lactis | CGMCC 1.2142 | CGMCC | 37 °C, MRS, Aerobic |
| Lactobacillus paracasei | CICC 20241 | CICC | 37 °C, MRS, Aerobic |
| Lactobacillus casei | CICC 20975 | CICC | 37 °C, MRS, Aerobic |
| Indicator Strains | Diameter of the Zone of Inhibition (mm) a,b | |
|---|---|---|
| JCM 1132 | CCFM 720 | |
| Gram-negative bacteria | ||
| ATCC 43893 | 8 | 8 |
| SL1344 | 8 | 8 |
| Gram-positive bacteria | ||
| S16 | 8 | 8 |
| S17 | 8 | 8 |
| S40 | 8 | 8 |
| W11 | 8 | 8 |
| W13 | 8 | 8 |
| W19 | 8 | 8 |
| S. aureus | 8 | 8 |
| E. faecalis | 8 | 8 |
| 9-5 | 8 | 8 |
| L103 | 8 | 8 |
| N5 | 8 | 8 |
| LGG | 8 | 8 |
| ZX5 | 8 | 8 |
| D2 | 8 | 8 |
| FFJND6-L5 | 8 | 8 |
| FHUBES1M18 | 8 | 8 |
| FHUBES1M24 | 8 | 8 |
| ATCC 19114 | 8 | 8 |
| ATCC 25175 | 8 | 8 |
| JCM 1557 | 11.3 ± 0.3 | 8 |
| CGMCC 1.2142 | 10.7 ± 0.3 | 8 |
| CICC 20241 | 8 | 8 |
| Treatment | Diameter of the Zone of Inhibition for CFS of JCM 1132 (mm) a,b |
|---|---|
| Without Protease | 11.3 ± 0.3 |
| Catalase | 11.0 ± 0.0 |
| Pepsin | 8 |
| Trypsin | 8 |
| Papain | 8 |
| Treatment | Diameter of the Zone of Inhibition for CFS of JCM 1132 (mm) a,b | Diameter of the Zone of Inhibition for CFS of CCFM 720 (mm) a,b |
|---|---|---|
| Without Heat | 11.3 ± 0.3 | 8 |
| 60 °C for 10 min | 10.8 ± 0.2 | 8 |
| 60 °C for 30 min | 10.5 ± 0.2 | 8 |
| 60 °C for 1 h | 10.5 ± 0.1 | 8 |
| 90 °C for 10 min | 10.3 ± 0.2 | 8 |
| 90 °C for 30 min | 10.3 ± 0.1 | 8 |
| 121 °C for 15 min | 10.3 ± 0.2 | 8 |
| Species | Strain | Generation Time | Adherence Index |
|---|---|---|---|
| L. acidophilus | JCM 1132 | 0.77 | 7.4 ± 0.5 |
| CCFM 720 | 0.77 | 8.1 ± 0.3 |
| Group | After Gavage for 3 Weeks (d21) a,b | After Withdraw for 1 Week (d28) a,b | ||||
|---|---|---|---|---|---|---|
| Simpson_1-D | Shannon_H | Chao-1 | Simpson_1-D | Shannon_H | Chao-1 | |
| CON | 0.96 ± 0.00 | 4.77 ± 0.13 | 15,560.50 ± 3660.38 | 0.95 ± 0.00 | 4.53 ± 0.19 | 16,100.00 ± 2001.66 |
| JCM 1132 | 0.96 ± 0.01 | 4.75 ± 0.25 | 13,123.33 ± 727.97 | 0.95 ± 0.01 | 4.63 ± 0.26 | 18,068.33 ± 4413.65 |
| CCFM 720 | 0.95 ± 0.02 | 4.81 ± 0.22 | 12,556.67 ± 1407.46 | 0.95 ± 0.01 | 4.72 ± 0.23 | 16,121.67 ± 3694.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Yu, Y.; Garcia-Gutierrez, E.; Jin, X.; He, Y.; Wang, L.; Tian, P.; Liu, Z.; Zhao, J.; Zhang, H.; et al. Lactobacillus acidophilus JCM 1132 Strain and Its Mutant with Different Bacteriocin-Producing Behaviour Have Various In Situ Effects on the Gut Microbiota of Healthy Mice. Microorganisms 2020, 8, 49. https://doi.org/10.3390/microorganisms8010049
Wang G, Yu Y, Garcia-Gutierrez E, Jin X, He Y, Wang L, Tian P, Liu Z, Zhao J, Zhang H, et al. Lactobacillus acidophilus JCM 1132 Strain and Its Mutant with Different Bacteriocin-Producing Behaviour Have Various In Situ Effects on the Gut Microbiota of Healthy Mice. Microorganisms. 2020; 8(1):49. https://doi.org/10.3390/microorganisms8010049
Chicago/Turabian StyleWang, Gang, Yunxia Yu, Enriqueta Garcia-Gutierrez, Xing Jin, Yufeng He, Linlin Wang, Peijun Tian, Zhenmin Liu, Jianxin Zhao, Hao Zhang, and et al. 2020. "Lactobacillus acidophilus JCM 1132 Strain and Its Mutant with Different Bacteriocin-Producing Behaviour Have Various In Situ Effects on the Gut Microbiota of Healthy Mice" Microorganisms 8, no. 1: 49. https://doi.org/10.3390/microorganisms8010049
APA StyleWang, G., Yu, Y., Garcia-Gutierrez, E., Jin, X., He, Y., Wang, L., Tian, P., Liu, Z., Zhao, J., Zhang, H., & Chen, W. (2020). Lactobacillus acidophilus JCM 1132 Strain and Its Mutant with Different Bacteriocin-Producing Behaviour Have Various In Situ Effects on the Gut Microbiota of Healthy Mice. Microorganisms, 8(1), 49. https://doi.org/10.3390/microorganisms8010049