Isolation and Characterization of Genes Responsible for Naphthalene Degradation from Thermophilic Naphthalene Degrader, Geobacillus sp. JF8
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Southern Blot Analyses
2.3. Cloning and DNA Sequencing
2.4. Enzymatic Assays
2.5. Purification of the Cis-Dihydrodiol Dehydrogenase
2.6. Electrophoresis
2.7. RNA Extraction and RT-PCR
2.8. Construction of an Expression Vector of nahAcAd and Its Biotransformation Assay
2.9. Nucleotide Sequence Accession Number
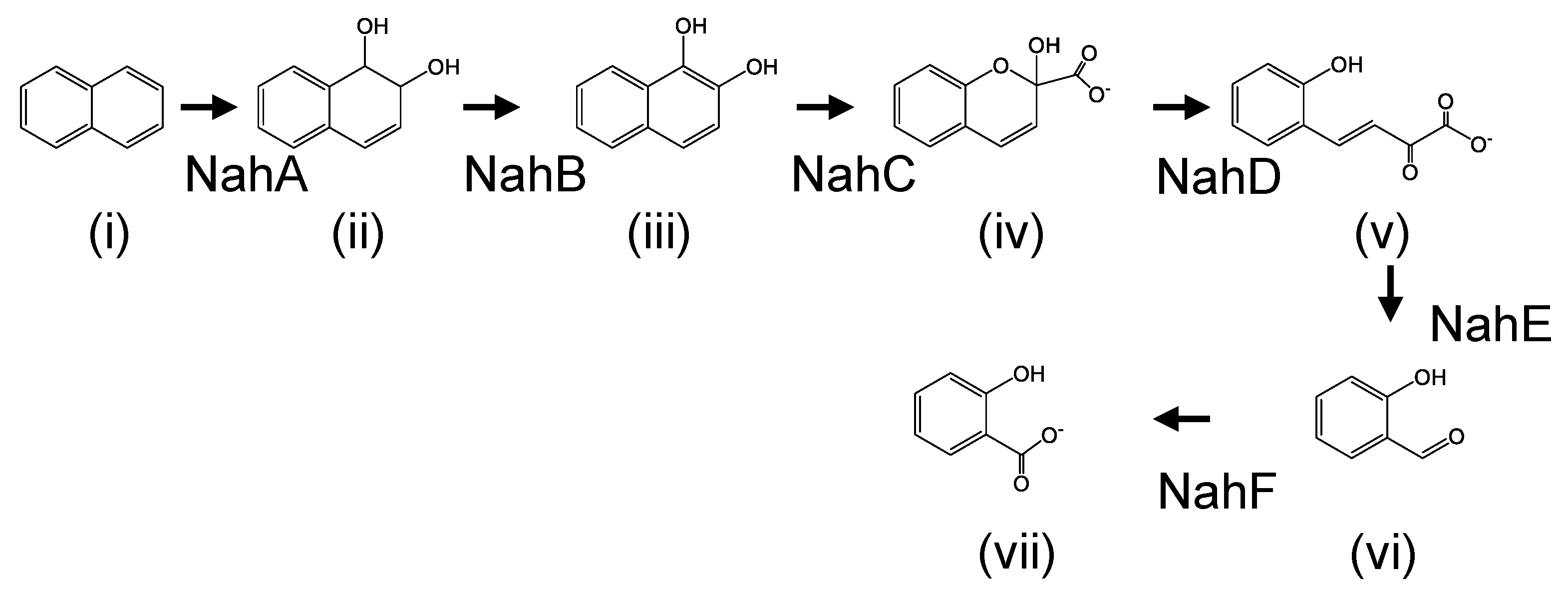
3. Results
3.1. Determination of N-terminal Amino Acid Sequence of Cis-Naphthalene Dihydrodiol Dehydrogenase
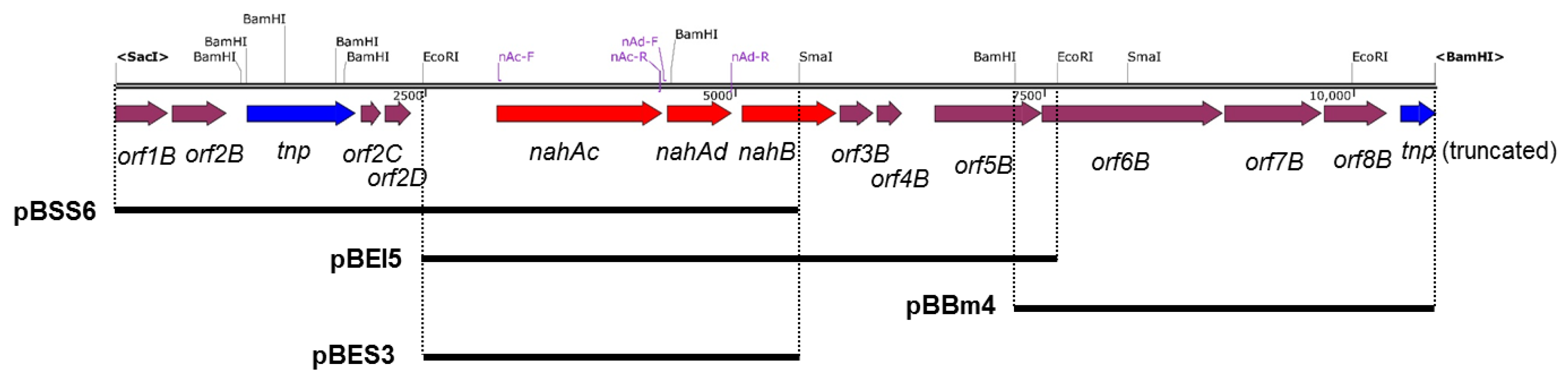
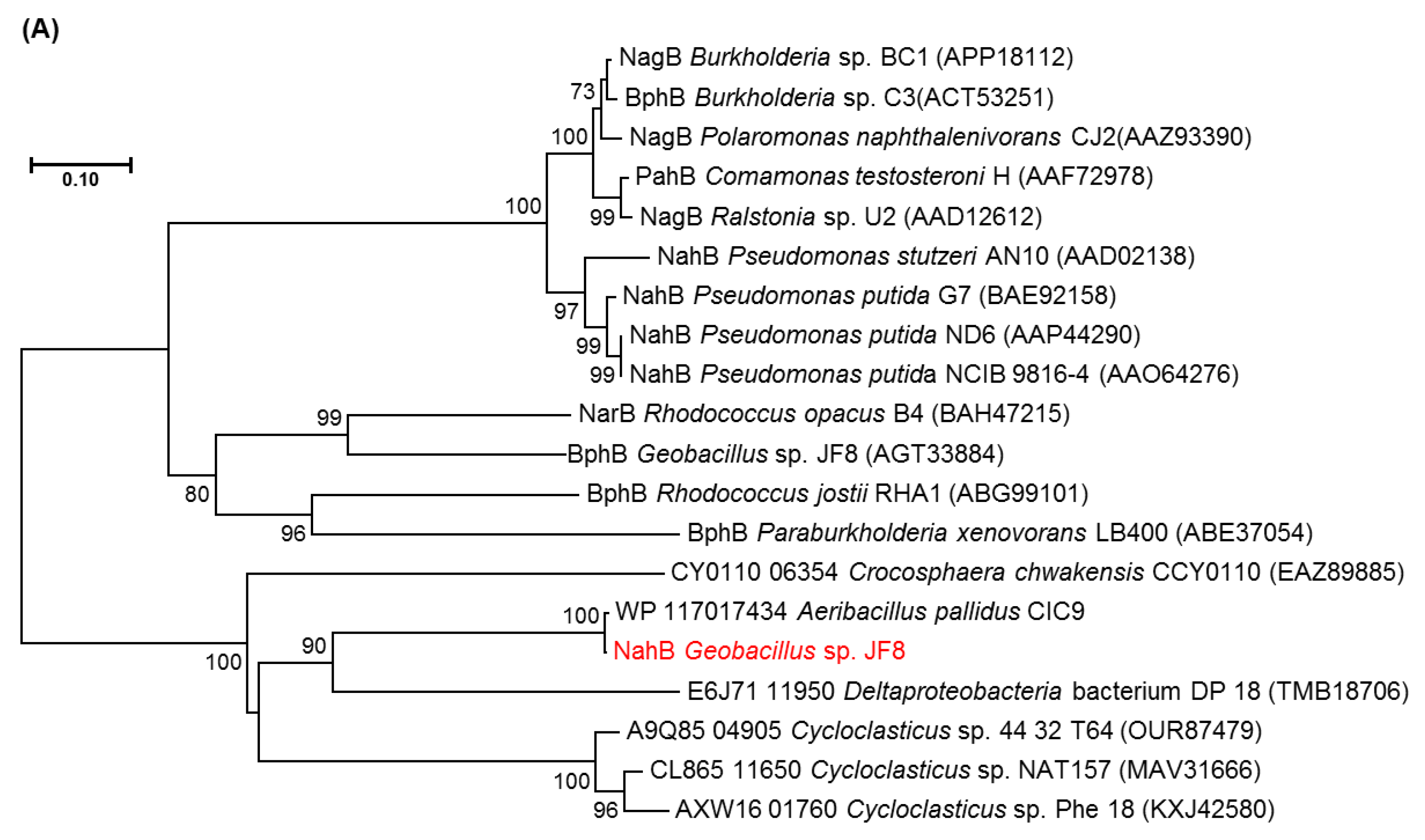
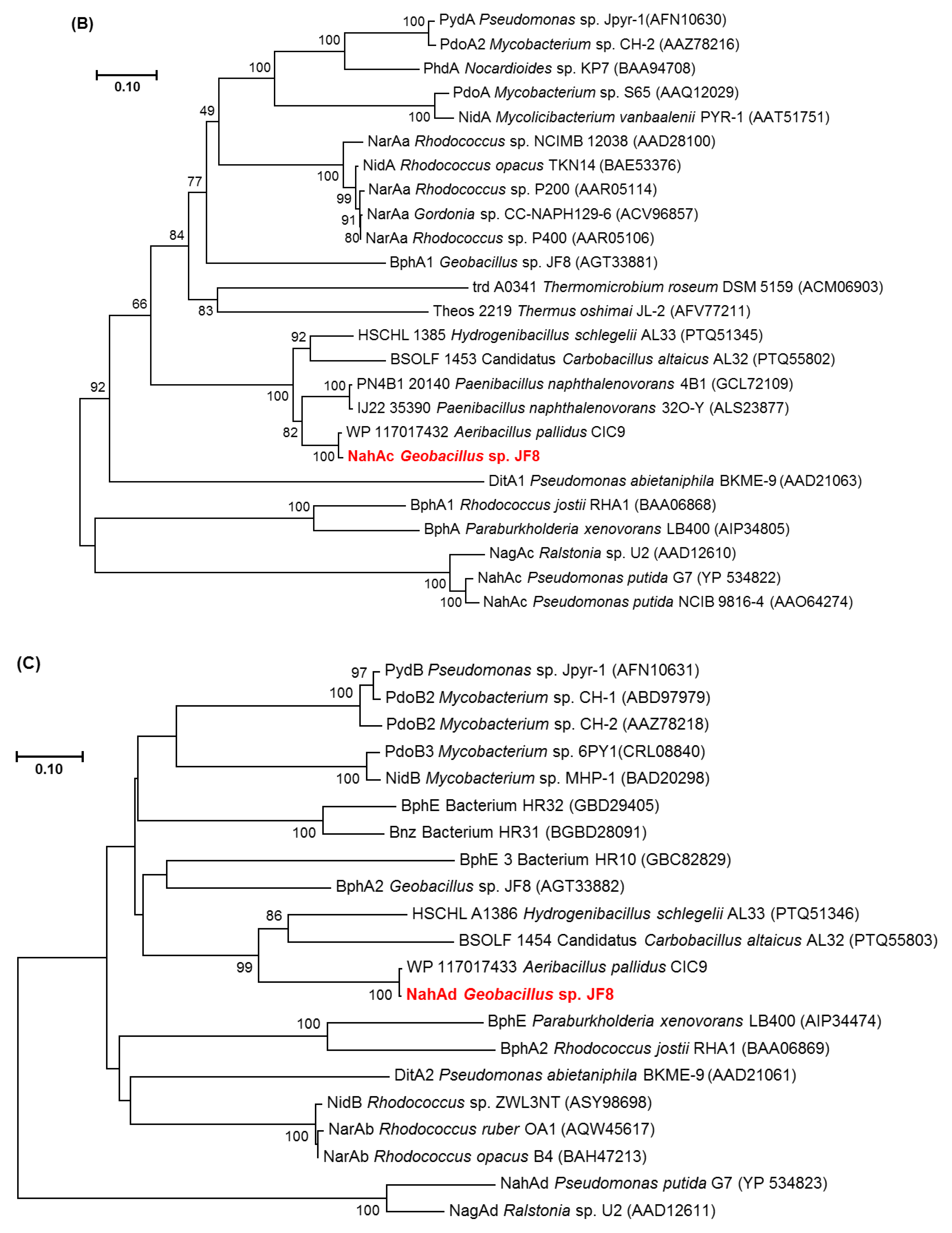
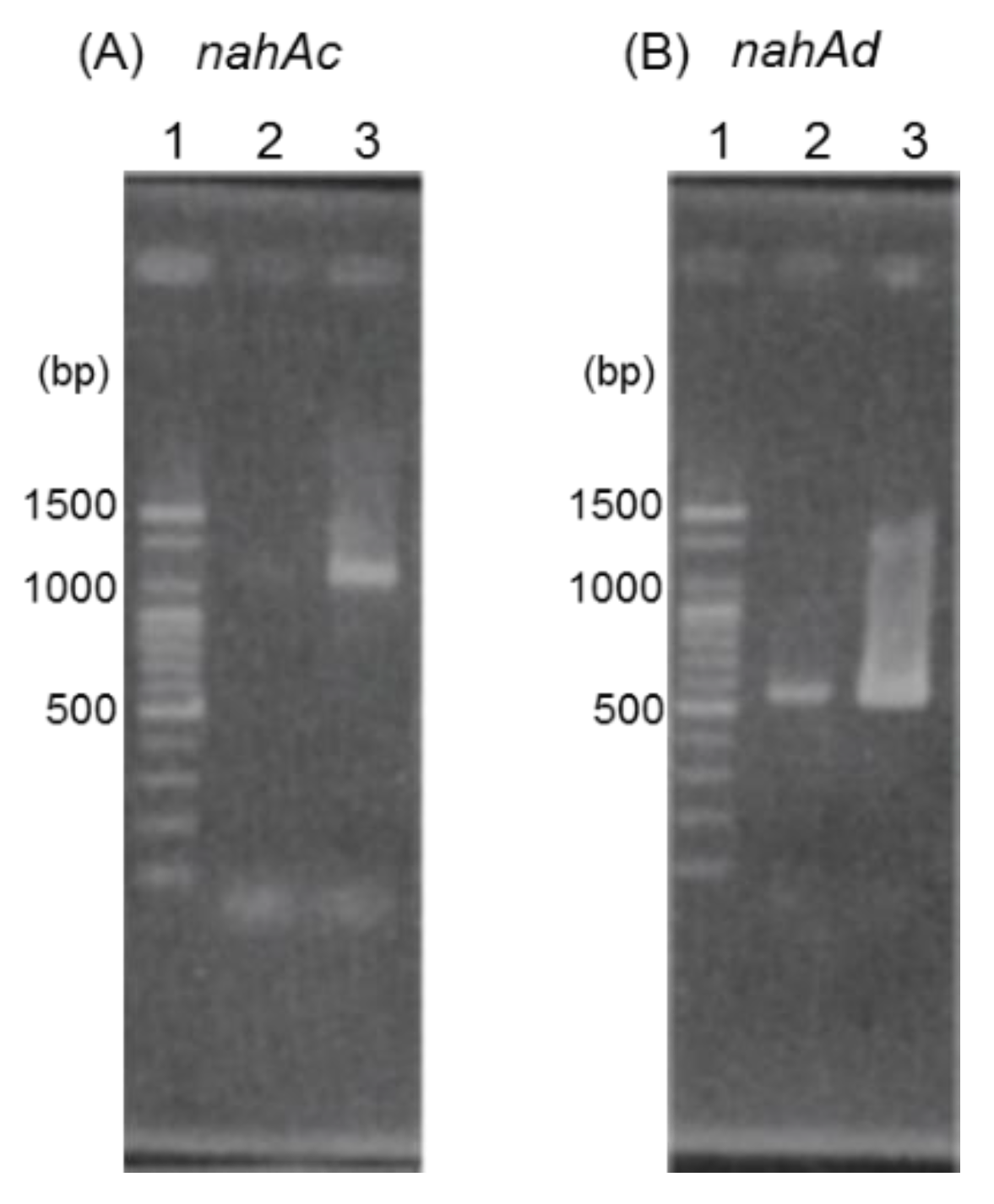
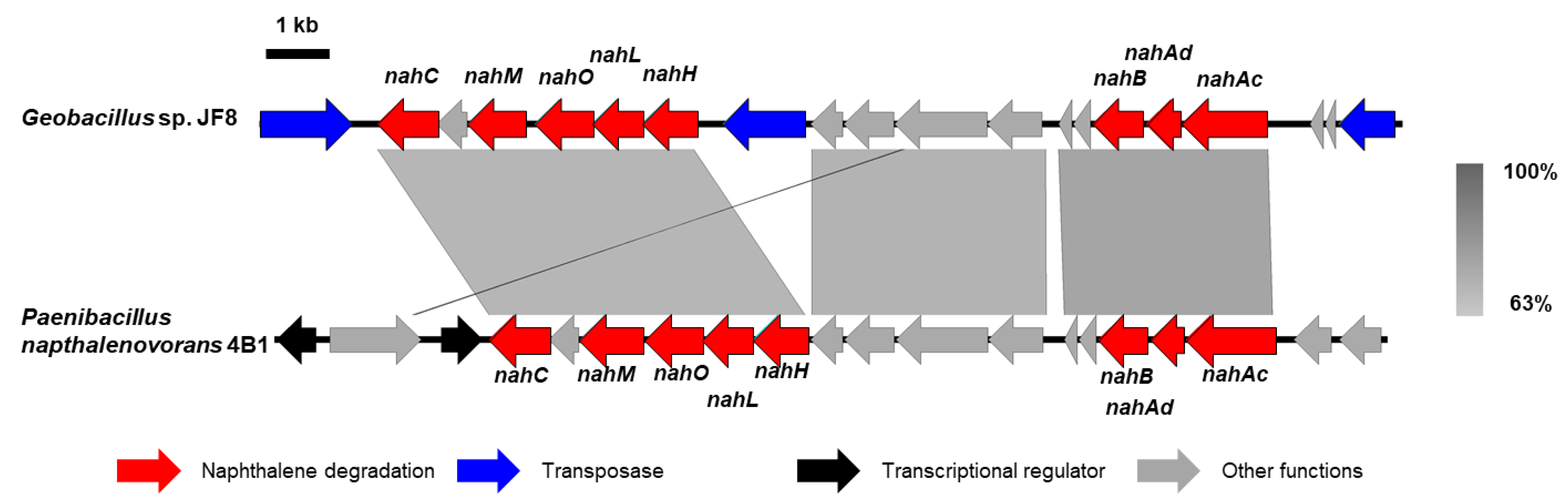
3.2. Cloning and Characterization of nah Genes
3.3. Gene Expression Analysis
3.4. Biotransformation Assay of nahAc and nahAd Gene Products
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Yen, K.M.; Gunsalus, I.C. Plasmid gene organization: Naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. USA 1982, 79, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Schell, M.A. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseudomonas putida. Proc. Natl. Acad. Sci. USA 1986, 83, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Harayama, S.; Rekik, M.; Wasserfallen, A.; Bairoch, A. Evolutionary relationships between catabolic pathways for aromatics: Conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol. Genet. Genom. 1987, 210, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.J.; Osslund, T.D.; Saunders, R.; Ensley, B.D.; Suggs, S.; Harcourt, A.; Suen, W.C.; Cruden, D.L.; Gibson, D.T.; Zylstra, G.J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 1993, 127, 31–37. [Google Scholar] [CrossRef]
- Cane, P.; Williams, P. A restriction map of naphthalene catabolic plasmid pWW60-1 and the location of some of its catabolic genes. Microbiology 1986, 132, 2919–2929. [Google Scholar] [CrossRef]
- Platt, A.; Shingler, V.; Taylor, S.C.; Williams, P.A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology 1995, 141, 2223–2233. [Google Scholar] [CrossRef]
- Schell, M.A.; Wender, P.E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J. Bacteriol. 1986, 166, 9–14. [Google Scholar] [CrossRef]
- Fuenmayor, S.L.; Wild, M.; Boyes, A.L.; Williams, P.A. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 1998, 180, 2522–2530. [Google Scholar]
- Zhou, N.Y.; Al-Dulayymi, J.; Baird, M.S.; Williams, P.A. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: A monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 2002, 184, 1547–1555. [Google Scholar] [CrossRef]
- Shimura, M.; Mukerjee-Dhar, G.; Kimbara, K.; Nagato, H.; Kiyohara, H.; Hatta, T. Isolation and characterization of a thermophilic Bacillus sp. JF8 capable of degrading polychlorinated biphenyls and naphthalene. FEMS Microbiol. Lett. 1999, 178, 87–93. [Google Scholar] [CrossRef]
- Annweiler, E.; Richnow, H.H.; Antranikian, G.; Hebenbrock, S.; Garms, C.; Franke, S.; Francke, W.; Michaelis, W. Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans. Appl. Environ. Microbiol. 2000, 66, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Bubinas, A.; Giedraitytė, G.; Kalėdienė, L.; Nivinskiene, O.; Butkiene, R. Degradation of naphthalene by thermophilic bacteria via a pathway, through protocatechuic acid. Cent. Eur. J. Biol. 2008, 3, 61–68. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Liu, J.; Li, R.; Shen, B. Isolation of a thermophilic bacterium, Geobacillus sp. SH-1, capable of degrading aliphatic hydrocarbons and naphthalene simultaneously, and identification of its naphthalene degrading pathway. Bioresour. Technol. 2012, 124, 83–89. [Google Scholar] [CrossRef]
- Miyazawa, D.; Mukerjee-Dhar, G.; Shimura, M.; Hatta, T.; Kimbara, K. Genes for Mn(ii)-dependent NahC and Fe(ii)-dependent NahH located in close proximity in the thermophilic naphthalene and PCB degrader, Bacillus sp. JF8: Cloning and characterization. Microbiology 2004, 150, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, N.; Iida, T.; Sawada, T.; Yamauchi, K.; Wang, Y.W.; Fukuda, M.; Kiyohara, H. Nucleotide sequences and characterization of genes encoding naphthalene upper pathway of Pseudomonas aeruginosa PAK1 and Pseudomonas putida OUS82. J. Biosci. Bioeng. 1999, 87, 721–731. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Patel, T.R.; Gibson, D.T. Purification and propeties of (plus)-cis-naphthalene dihydrodiol dehydrogenase of Pseudomonas putida. J. Bacteriol. 1974, 119, 879–888. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Di Gennaro, P.; Terreni, P.; Masi, G.; Botti, S.; De Ferra, F.; Bestetti, G. Identification and characterization of genes involved in naphthalene degradation in Rhodococcus opacus R7. Appl. Microbiol. Biotechnol. 2010, 87, 297–308. [Google Scholar] [CrossRef]
- Thanh, L.T.H.; Thi, T.V.N.; Shintani, M.; Moriuchi, R.; Dohra, H.; Loc, N.H.; Kimbara, K. Isolation and characterization of a moderate thermophilic Paenibacillus naphthalenovorans strain 4B1 capable of degrading dibenzofuran from dioxin-contaminated soil in Vietnam. J. Biosci. Bioeng. 2019, 128, 571–577. [Google Scholar] [CrossRef]
- Miñana-Galbis, D.; Pinzón, D.L.; Lorén, J.G.; Manresa, À.; Oliart-Ros, R.M. Reclassification of Geobacillus pallidus (Scholz et al. 1988) Banat et al. 2004 as Aeribacillus pallidus gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1600–1604. [Google Scholar] [CrossRef]
- Shintani, M.; Ohtsubo, Y.; Fukuda, K.; Hosoyama, A.; Ohji, S.; Yamazoe, A.; Fujita, N.; Nagata, Y.; Tsuda, M.; Hatta, T.; et al. Complete genome sequence of the thermophilic polychlorinated biphenyl degrader Geobacillus sp. strain JF8 (NBRC 109937). Genome Announc. 2014, 2, e01213-13. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.R.; Cammack, R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 1992, 46, 277–305. [Google Scholar] [CrossRef] [PubMed]
- Kauppi, B.; Lee, K.; Carredano, E.; Parales, R.E.; Gibson, D.T.; Eklund, H.; Ramaswamy, S. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 1998, 6, 571–586. [Google Scholar] [CrossRef]
- Khan, A.A.; Walia, S.K. Expression, localization, and functional analysis of polychlorinated biphenyl degradation genes cbpABCD of Pseudomonas putida. Appl. Environ. Microbiol. 1991, 57, 1325–1332. [Google Scholar]
- Kim, E.; Aversano, P.J.; Romine, M.F.; Schneider, R.P.; Zylstra, G.J. Homology between genes for aromatic hydrocarbon degradation in surface and deep-subsurface Sphingomonas strains. Appl. Environ. Microbiol. 1996, 62, 1467–1470. [Google Scholar]
- Treadway, S.L.; Yanagimachi, K.S.; Lankenau, E.; Lessard, P.A.; Stephanopoulos, G.; Sinskey, A.J. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 1999, 51, 786–793. [Google Scholar] [CrossRef]
- Khan, A.A.; Wang, R.F.; Cao, W.W.; Doerge, D.R.; Wennerstrom, D.; Cerniglia, C.E. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 3577–3585. [Google Scholar] [CrossRef]
- Wang, R.F.; Wennerstrom, D.; Cao, W.W.; Khan, A.A.; Cerniglia, C.E. Cloning, expression, and characterization of the katG gene, encoding catalase-peroxidase, from the polycyclic aromatic hydrocarbon-degrading bacterium Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2000, 66, 4300–4304. [Google Scholar] [CrossRef]
- Saito, A.; Iwabuchi, T.; Harayama, S. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: Expression in Escherichia coli. J. Bacteriol. 2000, 182, 2134–2141. [Google Scholar] [CrossRef]
- Kasuga, K.; Habe, H.; Chung, J.S.; Yoshida, T.; Nojiri, H.; Yamane, H.; Omori, T. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 2001, 283, 195–204. [Google Scholar] [CrossRef]
- Larkin, M.J.; Allen, C.C.; Kulakov, L.A.; Lipscomb, D.A. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 1999, 181, 6200–6204. [Google Scholar] [PubMed]
- Andreoni, V.; Bernasconi, S.; Colombo, M.; van Beilen, J.B.; Cavalca, L. Detection of genes for alkane and naphthalene catabolism in Rhodococcus sp. strain 1BN. Environ. Microbiol. 2000, 2, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Heitkamp, M.A.; Franklin, W.; Cerniglia, C.E. Microbial metabolism of polycyclic aromatic hydrocarbons: Isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 1988, 54, 2549–2555. [Google Scholar] [PubMed]
- Dean-Ross, D.; Moody, J.D.; Freeman, J.P.; Doerge, D.R.; Cerniglia, C.E. Metabolism of anthracene by a Rhodococcus species. FEMS Microbiol. Lett. 2001, 204, 205–211. [Google Scholar] [CrossRef]
- Boldrin, B.; Tiehm, A.; Fritzsche, C. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl. Environ. Microbiol. 1993, 59, 1927–1930. [Google Scholar]
- Schneider, J.; Grosser, R.; Jayasimhulu, K.; Xue, W.; Warshawsky, D. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 1996, 62, 13–19. [Google Scholar]
- Kanaly, R.A.; Harayama, S. Advances in the field of high-molecular-weight polycyclic aromatic hydrocarbon biodegradation by bacteria. Microb. Biotechnol. 2010, 3, 136–164. [Google Scholar] [CrossRef]
- Cebron, A.; Norini, M.P.; Beguiristain, T.; Leyval, C. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (pah-rhdalpha) genes from gram positive and gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 2008, 73, 148–159. [Google Scholar] [CrossRef]
- Mehetre, G.T.; Dastager, S.G.; Dharne, M.S. Biodegradation of mixed polycyclic aromatic hydrocarbons by pure and mixed cultures of biosurfactant producing thermophilic and thermo-tolerant bacteria. Sci. Total Environ. 2019, 679, 52–60. [Google Scholar] [CrossRef]
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H.; et al. Synergistic degradation of pyrene by five culturable bacteria in a mangrove sediment-derived bacterial consortium. J. Hazard. Mater. 2018, 342, 561–570. [Google Scholar] [CrossRef]
| Step | Volume | Total Protein | Total Activity | Specific Activity | Yield |
|---|---|---|---|---|---|
| (mL) | (mg) | (U) | (U/mg) | (%) | |
| Cell-free extract | 150 | 536 | 122 | 0.228 | 100 |
| DEAE Toyopearl 650M | 148 | 58 | 21.9 | 0.378 | 18 |
| Phenyl Sepharose ((NH4)2SO4-eluted) | 48 | 4 | 7.8 | 1.95 | 6.4 |
| Phenyl Sepharose (H2O-eluted) | 11 | 0.8 | 0.30 | 0.375 | 0.25 |
| MonoQ | 1.0 | 0.075 | 0.091 | 1.21 | 0.074 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyazawa, D.; Thanh, L.T.H.; Tani, A.; Shintani, M.; Loc, N.H.; Hatta, T.; Kimbara, K. Isolation and Characterization of Genes Responsible for Naphthalene Degradation from Thermophilic Naphthalene Degrader, Geobacillus sp. JF8. Microorganisms 2020, 8, 44. https://doi.org/10.3390/microorganisms8010044
Miyazawa D, Thanh LTH, Tani A, Shintani M, Loc NH, Hatta T, Kimbara K. Isolation and Characterization of Genes Responsible for Naphthalene Degradation from Thermophilic Naphthalene Degrader, Geobacillus sp. JF8. Microorganisms. 2020; 8(1):44. https://doi.org/10.3390/microorganisms8010044
Chicago/Turabian StyleMiyazawa, Daisuke, Le Thi Ha Thanh, Akio Tani, Masaki Shintani, Nguyen Hoang Loc, Takashi Hatta, and Kazuhide Kimbara. 2020. "Isolation and Characterization of Genes Responsible for Naphthalene Degradation from Thermophilic Naphthalene Degrader, Geobacillus sp. JF8" Microorganisms 8, no. 1: 44. https://doi.org/10.3390/microorganisms8010044
APA StyleMiyazawa, D., Thanh, L. T. H., Tani, A., Shintani, M., Loc, N. H., Hatta, T., & Kimbara, K. (2020). Isolation and Characterization of Genes Responsible for Naphthalene Degradation from Thermophilic Naphthalene Degrader, Geobacillus sp. JF8. Microorganisms, 8(1), 44. https://doi.org/10.3390/microorganisms8010044





