Biotransformation of Monocyclic Phenolic Compounds by Bacillus licheniformis TAB7
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Chemicals and Reagents
2.3. Utilization of Phenolic Compounds as Sole Carbon Sources
2.4. Biotransformation Assays
2.5. Analytical Procedure
2.5.1. Sample Preparation for HPLC Analysis
2.5.2. HPLC Analysis
2.5.3. LC-MS /MS Analysis of Ferulate and Caffeate Degradation Products
3. Results and Discussion
3.1. Ability to Utilize Phenolic Compounds as Sole Carbon Sources
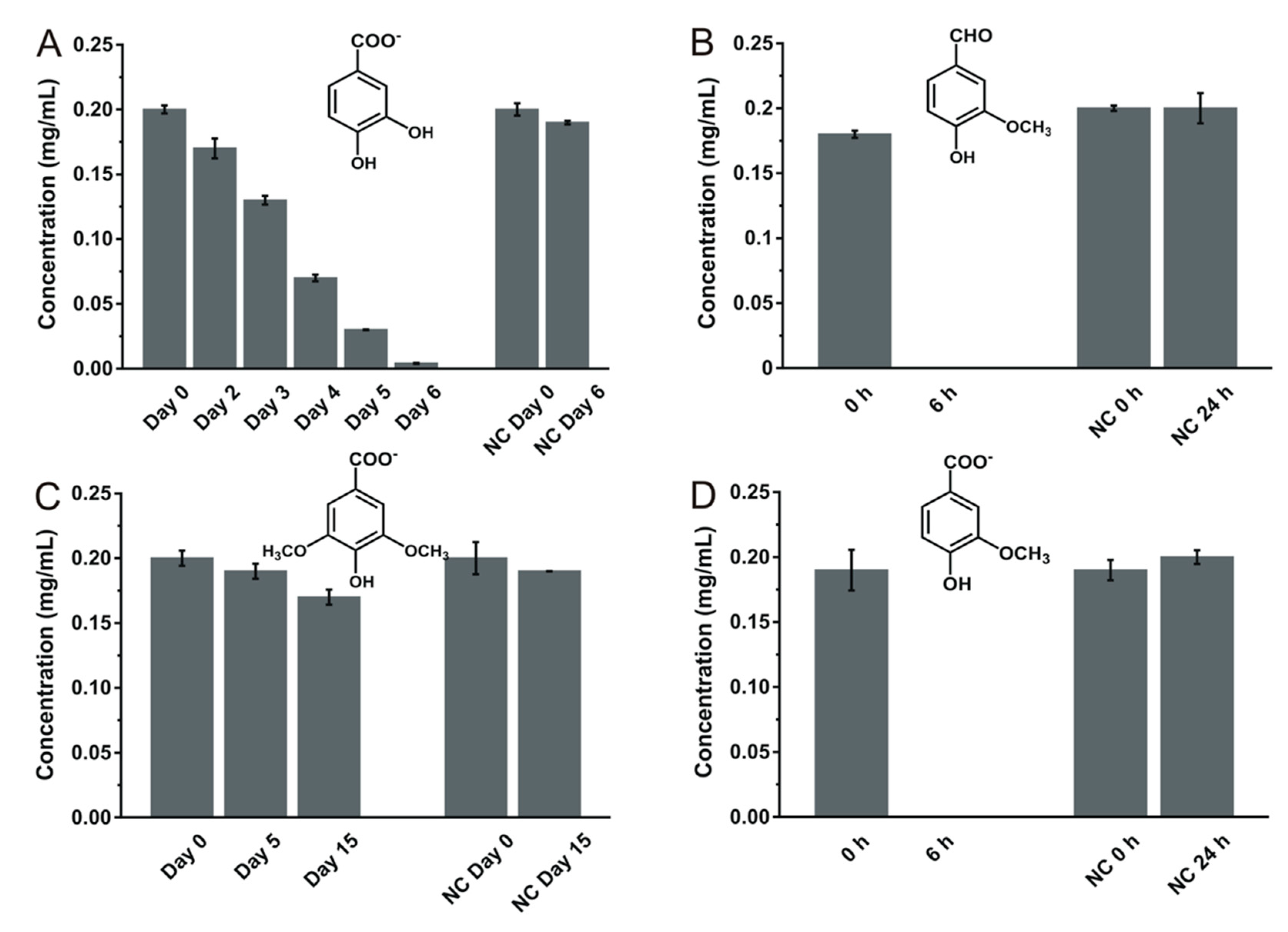
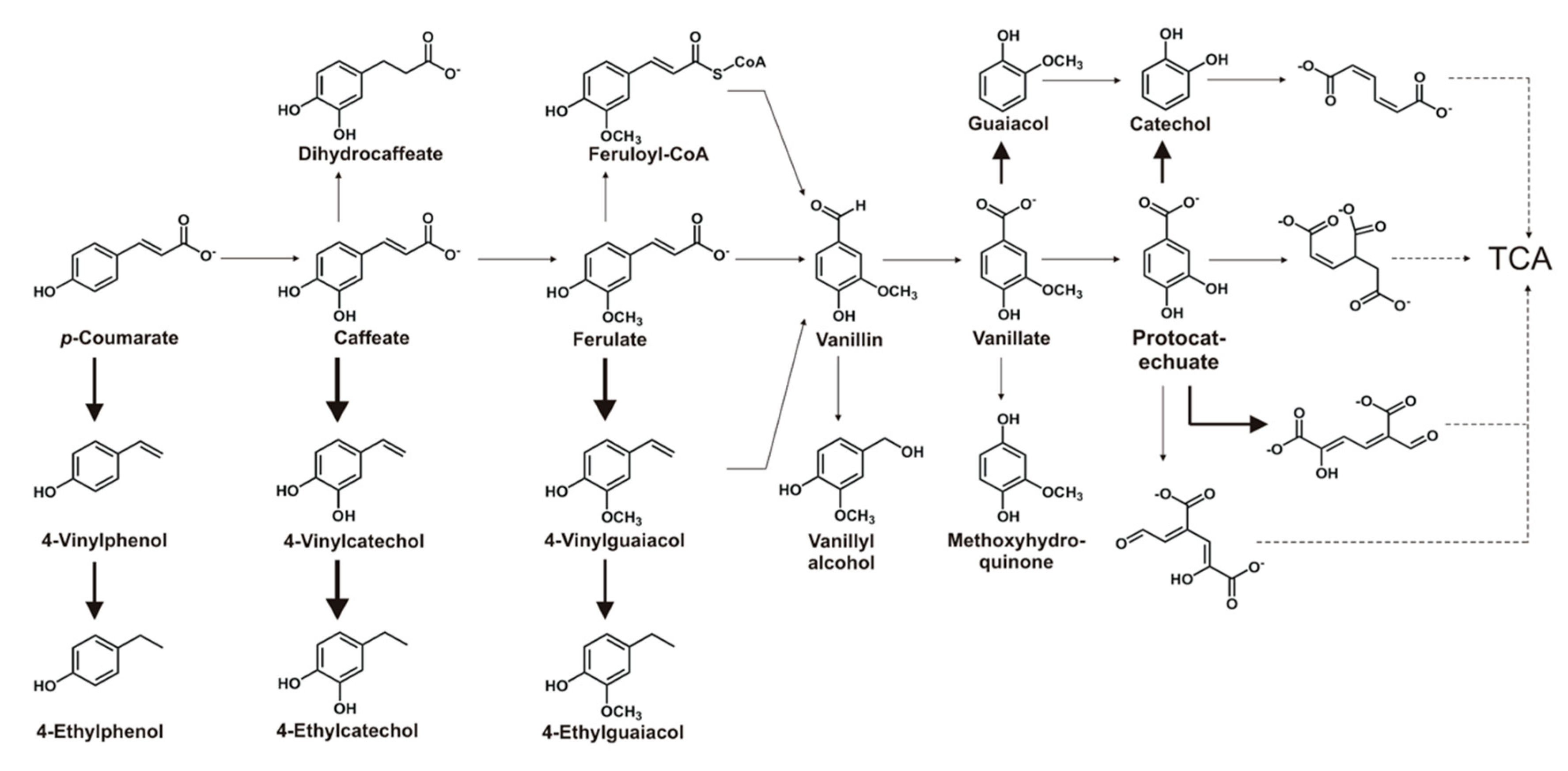
3.2. Transformation Ability for Phenolic Compounds
3.3. Identification of Intermediates/Products by LC-MS/MS
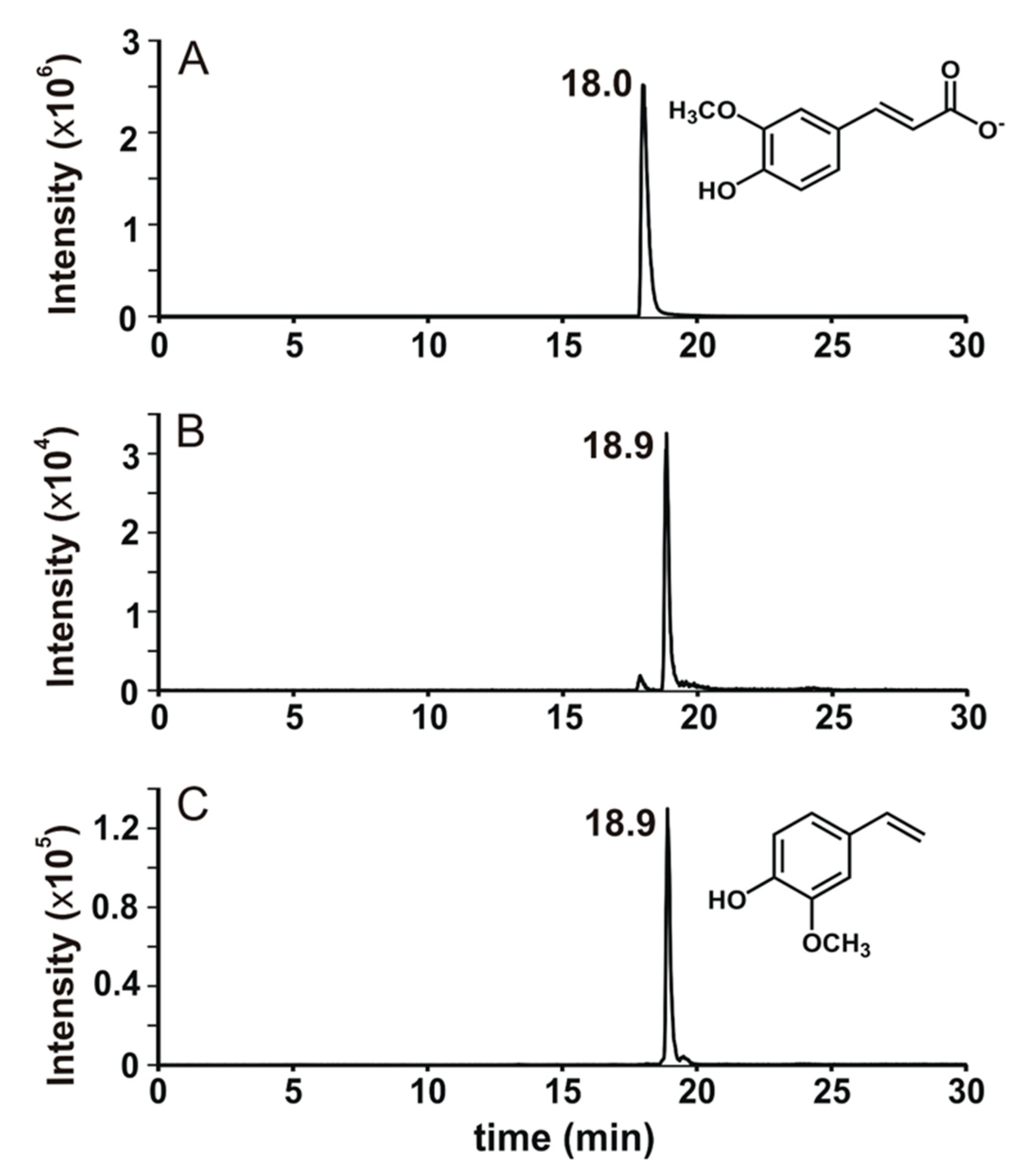
3.3.1. Identification of the Ferulate Biotransformation Product
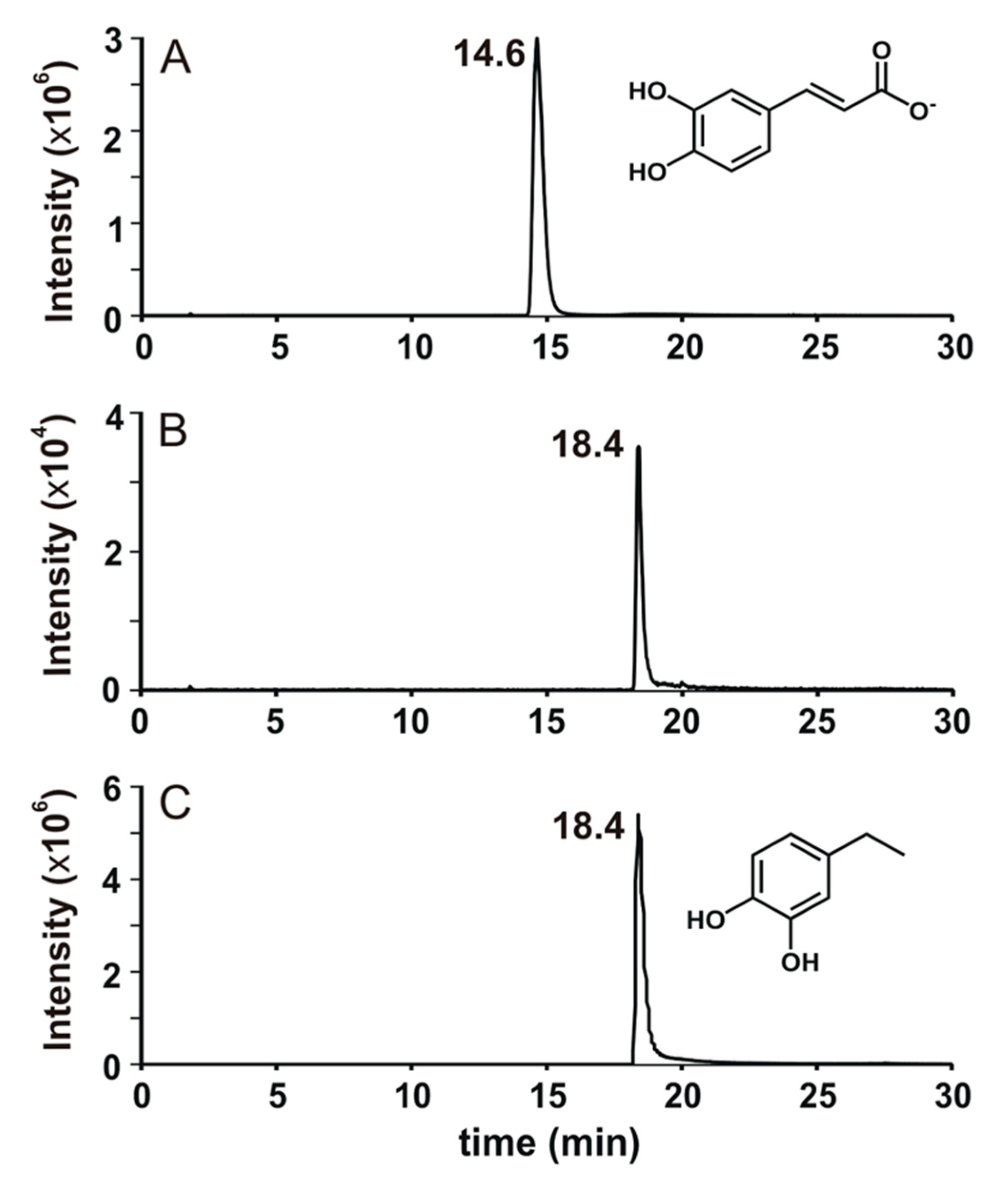
3.3.2. Identification of Caffeate Biotransformation Product
3.4. Phenolic Compounds Degradative Genes in TAB7
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kimura, T. Aerobic Composting of Livestock Manure by Thermophile and Enzymes. J. Environ. Biotechnol 2015, 15, 23–28. [Google Scholar]
- Toyota, Menicon Develop Advanced Composting Process. Available online: https://global.toyota/en/detail/274100 (accessed on 11 November 2019).
- Mendonca, E.; Martins, A.; Anselmo, A.M. Biodegradation of natural phenolic compounds as single and mixed substrates by Fusarium flocciferum. Electron. J. Biotechnol. 2004, 7, 30–37. [Google Scholar] [CrossRef][Green Version]
- Li, Z.H.; Wang, Q.; Ruan, X.; Pan, C.D.; Jiang, D.A. Phenolics and Plant Allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef] [PubMed]
- Kanchan, S.D.; Jayachandra. Allelopathic effects of Parthenium hysterophorus L. part IV. Identification of inhibitors. Plant Soil. 1980, 55, 67–75. [Google Scholar] [CrossRef]
- Patterson, D.T. Effects of allelopathic chemicals on growth and physiological responses of soybean (Glycine max). Weed Sci. 1981, 29, 53–59. [Google Scholar] [CrossRef]
- Blum, U. Allelopathic interactions involving phenolic acids. J. Nematol. 1996, 28, 259–267. [Google Scholar] [PubMed]
- Blum, U. Fate of Phenolic Allelochemicals in Soils—The Role of Soil and Rhizosphere Microorganisms. In Allelopathy: chemistry and mode of action of allelochemicals; Macías, F.A., Galindo, J.C.G., Molinillo, J.M.G., Cutler, H.G., Eds.; CRC Press: New York, NY, USA, 2005; pp. 57–72. [Google Scholar]
- Jilani, G.; Mahmood, S.; Chaudhry, A.N.; Hassan, I.; Akram, M. Allelochemicals: Sources, toxicity and microbial transformation in the soil—A review. Ann. Microbiol. 2008, 58, 351–357. [Google Scholar] [CrossRef]
- Mpofu, E.; Vejarano, F.; Suzuki-Minakuchi, C.; Ohtsubo, Y.; Tsuda, M.; Chakraborty, J.; Nakajima, M.; Okada, K.; Tada, N.; Kimura, T.; et al. Complete genome sequence of Bacillus licheniformis TAB7, a compost deodorizing strain with potential for plant-growth promotion. Microbiol. Resour. Announc. 2018, 8, e01659-18. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3, p. A2.2. [Google Scholar]
- Beek, S.; Priest, F.G. Decarboxylation of Substituted Cinnamic Acids by Lactic Acid Bacteria Isolated during Malt Whisky Fermentation. Appl. Environ. Microbiol. 2000, 66, 5322–5328. [Google Scholar] [CrossRef]
- Karmakar, B.; Vohra, R.M.; Nandanwar, H.; Sharma, P.; Gupta, K.G.; Sobti, R.C. Rapid degradation of ferulic acid via 4-vinylguaiacol and vanillin by a newly isolated strain of Bacillus coagulans. J. Biotechnol. 2000, 80, 195–202. [Google Scholar] [CrossRef]
- Lee, I.Y.; Volm, T.G.; Rosazza, J.P.N. Decarboxylation of ferulic acid to 4-vinylguaiacol by Bacillus pumilus in aqueous-organic solvent two-phase systems. Enzyme Microb. Technol. 1998, 23, 261–266. [Google Scholar] [CrossRef]
- Li, X.M.; Yang, J.K.; Li, X.; Gu, W.; Huang, J.W.; Zhang, K.Q. The metabolism of ferulic acid via 4-vinylguaiacol to vanillin by Enterobacter sp. Px6-4 isolated from Vanilla root. Process Biochem. 2008, 43, 1132–1137. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E.; Sudheesh, S. Rapid conversion of ferulic acid to 4-vinyl guaiacol and vanillin metabolites by Debaryomyces hansenii. J. Mol. Catal. B Enzym. 2007, 44, 48–52. [Google Scholar] [CrossRef]
- Koseki, T.; Ito, Y.; Furuse, S.; Ito, K.; Iwano, K. Conversion of ferulic acid into 4-vinylguaiacol, vanillin and vanillic acid in model solutions of Shochu. J. Ferment Bioeng. 1996, 82, 46–50. [Google Scholar] [CrossRef]
- Achterholt, S.; Priefert, H.; Steinbüchel, A. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 2000, 54, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yu, H.N.; Wu, Y.F.; Liu, X.Y.; Cheng, A.X.; Lou, H.X. Cloning and functional characterization of a phenolic acid decarboxylase from the liverwort, Conocephalum japonicum. Biochem. Biophys. Res. Commun. 2016, 481, 239–244. [Google Scholar] [CrossRef]
- Jones, C.D.; Woods, K.E.; Setzer, W.N. A chemical ecological investigation of the allelopathic potential of Lamium amplexicaule and Lamium Purpureum. Open Ecol. J. 2012, 2, 167–177. [Google Scholar] [CrossRef]
- Khanh, T.D.; Cong, L.C.; Xuan, T.D.; Lee, S.J.; Kong, D.S.; Chung, M. Weed-Suppressing Potential of Dodder (Cuscuta hygrophilae) and its Phytotoxic Constituents. Weed Sci. 2008, 56, 119–127. [Google Scholar] [CrossRef]
- Formisano, C.; Rigano, D.; Senatore, F.; De Feo, V.; Bruno, M.; Rosselli, S. Composition and allelopathic effect of essential oils of two thistles: Cirsium creticum (Lam.) D.’Urv. ssp. triumfetti (Lacaita) Werner and Carduus nutans L. J. Plant Interact. 2007, 2, 115–120. [Google Scholar] [CrossRef]
- Senger, D.R.; Li, D.; Jaminet, S.C.; Cao, S. Activation of the Nrf2 Cell Defense Pathway by Ancient Foods: Disease Prevention by Important Molecules and Microbes Lost from the Modern Western Diet. PLoS ONE 2016, 11, e0148042. [Google Scholar] [CrossRef]
- Buron, N.; Guichard, H.; Coton, E.; Ledauphin, J.; Barillier, D. Evidence of 4-ethylcatechol as one of the main phenolic off-flavor markers in French ciders. Food Chem. 2011, 125, 542–548. [Google Scholar] [CrossRef]
- Peppercorn, M.A.; Goldman, P. Caffeic Acid Metabolism by Bacteria of the Human Gastrointestinal Tract. J. Bacteriol. 1971, 108, 996–1000. [Google Scholar]
- Santamaría, L.; Reverón, I.; Felipe, F.L.; Rivas, B.; Muñoz, R. Ethylphenols formation by Lactobacillus plantarum: Identification of the enzyme involved in the reduction of vinylphenols. Appl. Environ. Microbiol. 2018, 84, e01064-18. [Google Scholar] [CrossRef] [PubMed]
- Tchobanov, I.; Gal, L.; Guilloux-Benatier, M.; Remize, F.; Nardi, T.; Guzzo, J.; Serpaggi, V.; Alexandre, H. Partial vinylphenol reductase purification and characterization from Brettanomyces Bruxellensis. Microbiol. Lett. 2008, 284, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Godoy, L.; Martínez, C.; Carrasco, N.; Ganga, M.A. Purification and characterization of a p-coumarate decarboxylase and a vinylphenol reductase from Brettanomyces. Int. J. Food Microbiol. 2008, 127, 6–11. [Google Scholar] [CrossRef]
- Harris, V.; Ford, C.M.; Jiranek, V.; Grbin, P.R. Survey of enzyme activity responsible for phenolic off-flavor production by Dekkera and Brettanomyces yeast. Appl. Microbiol. Biotechnol. 2009, 81, 1117–1127. [Google Scholar] [CrossRef]
- Iwagami, S.G.; Yang, K.; Davies, J. Characterization of the Protocatechuic Acid Catabolic Gene Cluster from Streptomyces sp. strain 2065. Appl. Environ. Microbiol. 2000, 66, 1499–1508. [Google Scholar] [CrossRef]
- Kasai, D.; Fujinami, T.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009, 191, 6758–6768. [Google Scholar] [CrossRef]
- Fujisawa, H.; Hayaishi, O. Protocatechuate 3,4-dioxygenase. I. Crystallization and characterization. J. Biol. Chem. 1968, 243, 2673–2681. [Google Scholar]
- Kamimura, N.; Aoyama, T.; Yoshida, R.; Takahashi, K.; Kasai, D.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Characterization of the protocatechuate 4,5-cleavage pathway operon in comamonas sp. strain e6 and discovery of a novel pathway gene. Appl. Environ. Microbiol. 2010, 76, 8093–8101. [Google Scholar] [CrossRef]
- Noda, Y.; Nishikawa, S.; Shiozuka, K.I.; Kadokura, H.; Nakajima, H.; Yoda, K.; Katayama, Y.; Morohoshi, N.; Haraguchi, T.; Yamasaki, M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 1990, 172, 2704–2709. [Google Scholar] [CrossRef] [PubMed]
- Wolgel, S.A.; Dege, J.A.Y.E.; Perkins-olson, P.E.; Juarez-garcia, C.H.; Crawford, R.L.; Munck, E.; Lipscomb, J.D. Purification and Characterization of Protocatechuate Extradiol Catecholic Dioxygenase. J. Bacteriol. 1993, 175, 4414–4426. [Google Scholar] [CrossRef] [PubMed]
- Granato, T.M.; Romano, D.; Vigentini, I.; Foschino, R.C.; Monti, D.; Mamone, G.; Ferranti, P.; Nitride, C.; Iametti, S.; Bonomi, F.; et al. New insights on the features of the vinyl phenol reductase from the wine-spoilage yeast Dekkera/Brettanomyces bruxellensis. Ann. Microbiol. 2015, 65, 321–329. [Google Scholar] [CrossRef]
- Nogales, J.; Canales, Á.; Jiménez-Barbero, J.; Serra, B.; Pingarrón, J.M.; García, J.L.; Díaz, E. Unravelling the gallic acid degradation pathway in bacteria: The gal cluster from Pseudomonas putida. Mol. Microbiol. 2011, 79, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Kasai, D.; Masai, E.; Miyauchi, K.; Katayama, Y.; Fukuda, M. Characterization of the 3-O-Methylgallate Dioxygenase Gene and Evidence of Multiple 3-O-Methylgallate Catabolic Pathways in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 2004, 186, 4951–4959. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mpofu, E.; Chakraborty, J.; Suzuki-Minakuchi, C.; Okada, K.; Kimura, T.; Nojiri, H. Biotransformation of Monocyclic Phenolic Compounds by Bacillus licheniformis TAB7. Microorganisms 2020, 8, 26. https://doi.org/10.3390/microorganisms8010026
Mpofu E, Chakraborty J, Suzuki-Minakuchi C, Okada K, Kimura T, Nojiri H. Biotransformation of Monocyclic Phenolic Compounds by Bacillus licheniformis TAB7. Microorganisms. 2020; 8(1):26. https://doi.org/10.3390/microorganisms8010026
Chicago/Turabian StyleMpofu, Enock, Joydeep Chakraborty, Chiho Suzuki-Minakuchi, Kazunori Okada, Toshiaki Kimura, and Hideaki Nojiri. 2020. "Biotransformation of Monocyclic Phenolic Compounds by Bacillus licheniformis TAB7" Microorganisms 8, no. 1: 26. https://doi.org/10.3390/microorganisms8010026
APA StyleMpofu, E., Chakraborty, J., Suzuki-Minakuchi, C., Okada, K., Kimura, T., & Nojiri, H. (2020). Biotransformation of Monocyclic Phenolic Compounds by Bacillus licheniformis TAB7. Microorganisms, 8(1), 26. https://doi.org/10.3390/microorganisms8010026



