Molecular Biomarkers and Influential Factors of Denitrification in a Full-Scale Biological Nitrogen Removal Plant
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Wastewater Sample Collection
2.2. Bacterial Pure Cultures, Culture Media, and Growth Conditions
2.3. DNA Extraction
2.4. PCR and qPCR Assays
2.5. Analysis of PCR Products, PCR Product Purification, and DNA Sequencing
2.6. Melt Curves
2.7. Standards for Real-Time PCR
2.8. Real-Time PCR Result Calculation
2.9. Physical and Chemical Analysis
2.10. Statistical Analysis
3. Results
3.1. Denitrifying Bacteria and Denitrifying Genes in this Study
3.2. Operational Conditions, Wastewater Characteristics, and Bioreactor Performance
3.3. Temporal Dynamics of Denitrifying Bacteria and Functional Genes
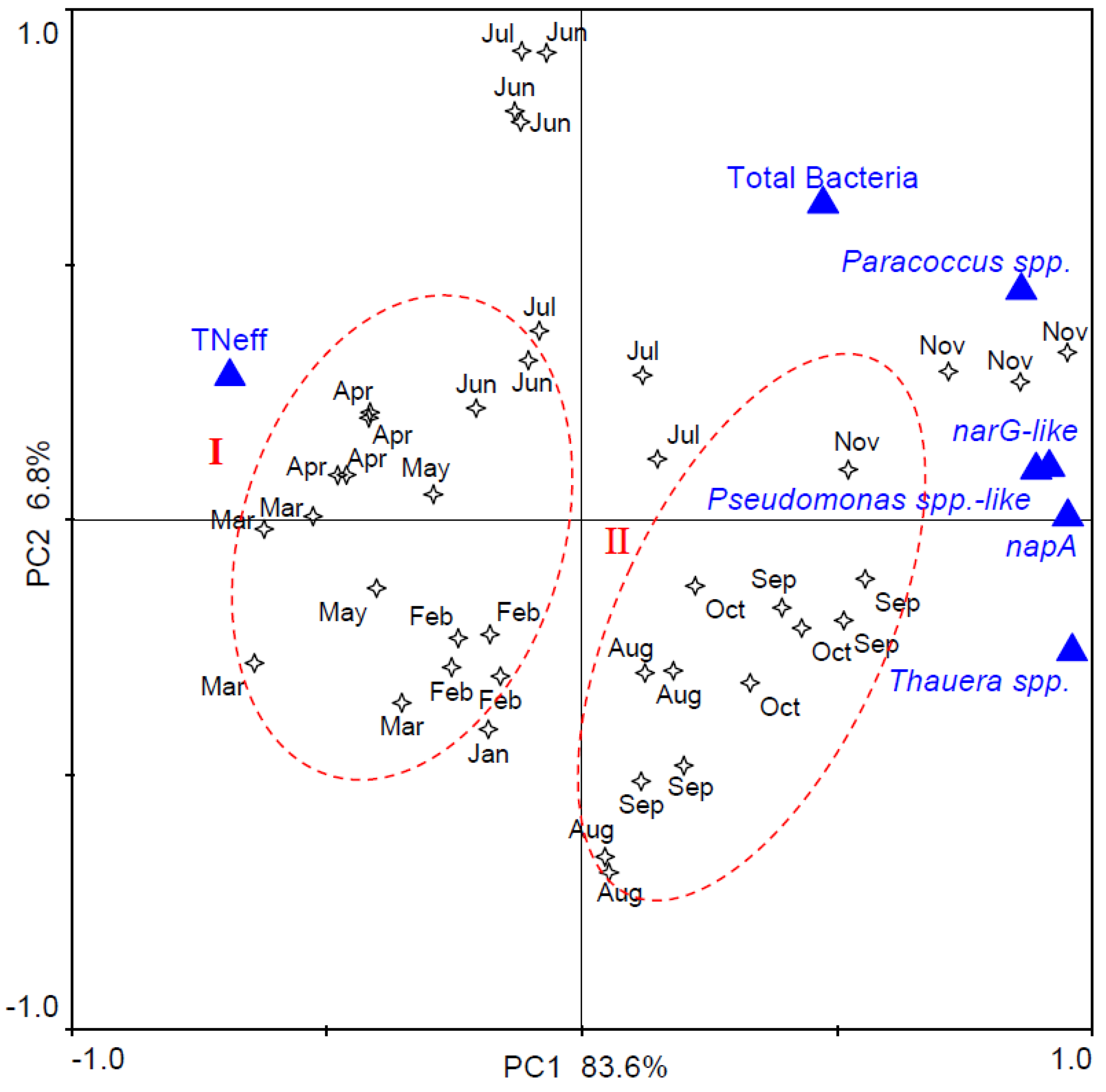
3.4. Correlation between Denitrifier Abundance, Wastewater Characteristics, and Operational Conditions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lu, H.J.; Chandran, K.; Stensel, D. Microbial Ecology of Denitrification in Biological Wastewater Treatment. Water Res. 2014, 64, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Bitton, G. Wastewater Microbiology, 4th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Tchobanoglous, G.; Burton, F.L.; Stensel, H.D. Wastewater Engineering: Treatment and Reuse, 4th ed.; McGraw-Hill: Boston, ME, USA, 2003. [Google Scholar]
- Rittmann, B.E.; Laspidou, C.S.; Flax, J.; Stahl, D.A.; Urbain, V.; Harduin, H.; van der Waarde, J.J.; Geurkink, B.; Henssen, M.J.C.; Brouwer, H.; et al. Molecular and Modeling Analyses of the Structure and Function of Nitrifying Activated Sludge. Water Sci. Technol. 1999, 39, 51–59. [Google Scholar] [CrossRef]
- Lu, H.J.; Nuruzzaman, F.; Ravindhar, J.; Chandran, K. Alcohol Dehydrogenase Expression as a Biomarker of Denitrification Activity in Activated Sludge using Methanol and Glycerol as Electron Donors. Environ. Microbiol. 2011, 13, 2930–2938. [Google Scholar] [CrossRef] [PubMed]
- Enwall, K.; Philippot, L.; Hallin, S. Activity and Composition of the Denitrifying Bacterial Community Respond Differently to Long-Term Fertilization. Appl. Environ. Microbiol. 2005, 71, 8335–8343. [Google Scholar] [CrossRef]
- Shapleigh, J.P. The Denitrifying Prokaryotes. In The Prokaryotes a Handbook on the Biology of Bacteria: Ecophysiology and Biochemistry; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 2, pp. 769–792. [Google Scholar]
- Chakravarthy, S.S.; Pande, S.; Kapoor, A.; Nerurkar, A.S. Comparison of Denitrification between Paracoccus sp. and Diaphorobacter sp. Appl. Biochem. Biotech. 2011, 165, 260–269. [Google Scholar] [CrossRef]
- Thomsen, T.R.; Kong, Y.; Nielsen, P.H. Ecophysiology of Abundant Denitrifying Bacteria in Activated Sludge. FEMS Microbiol. Ecol. 2007, 60, 370–382. [Google Scholar] [CrossRef]
- Heylen, K.; Vanparys, B.; Wittebolle, L.; Verstraete, W.; Boon, N.; De Vos, P. Cultivation of Denitrifying Bacteria: Optimization of Isolation Conditions and Diversity Study. Appl. Environ. Microbiol. 2006, 72, 2637–2643. [Google Scholar] [CrossRef]
- Neef, A.; Zaglauer, A.; Meier, H.; Amann, R.; Lemmer, H.; Schleifer, K.H. Population Analysis in a Denitrifying Sand Filter: Conventional and in Situ Identification of Paracoccus spp. in Methanol-Fed biofilms. Appl. Environ. Microbiol. 1996, 62, 4329–4339. [Google Scholar]
- Philippot, L.; Hallin, S. Finding the Missing Link Between Diversity and Activity using Denitrifying Bacteria as a Model Functional Community. Curr. Opin. Microbiol. 2005, 8, 234–239. [Google Scholar] [CrossRef]
- Philippot, L.; Piutti, S.; Martin-Laurent, F.; Hallet, S.; Germon, J.C. Molecular Analysis of the Nitrate-Reducing Community from Unplanted and Maize-Planted Soils. Appl. Environ. Microbiol. 2002, 68, 6121–6128. [Google Scholar] [CrossRef]
- Gedalanga, P.B.; Olson, B.H. Development of a Quantitative PCR Method to Differentiate between Viable and Nonviable Bacteria in Environmental Water Samples. Appl. Microbiol. Biotechnol. 2009, 82, 587–596. [Google Scholar] [CrossRef]
- Yu, Z.T.; Mohn, W.W. Killing Two Birds with One Stone: Simultaneous Extraction of DNA and RNA from Activated Sludge Biomass. Can. J. Microbiol. 1999, 45, 269–272. [Google Scholar] [CrossRef]
- Asvapathanagul, P. Application of Molecular Tools to Understand Foaming and Bulking Filamentous Bacteria in Activated Sludge. Ph.D. Thesis, University of California, Irvine, CA, USA, 2011. [Google Scholar]
- Asvapathanagul, P.; Olson, B.H.; Gedalanga, P.B.; Hashemi, A.; Huang, Z.H.; La, J. Identification and Quantification of Thiothrix eikelboomii using qPCR for Early Detection of Bulking Incidents in a Full-Scale Water Reclamation Plant. Appl. Microbiol. Biotechnol. 2015, 99, 4045–4057. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y. Fine-Scale Microbial Analysis on Denitrifying Bacteria for the Improved Control of Wastewater Treatment. Ph.D. Thesis, University of California, Irvine, CA, USA, 2012. [Google Scholar]
- Wang, T. Understanding of Nitrifying and Denitrifying Bacterial Population Dynamics in an Activated Sludge Process. Ph.D. Thesis, University of California at Irvine, Irvine, CA, USA, 2014. [Google Scholar]
- Lane, D.J. Nucleic Acid Techniques in Bacterial Systematics; John Wiley & Sons: Hoboken, NJ, USA, 1991. [Google Scholar]
- Ferris, M.; Muyzer, G.; Ward, D.M. Denaturing Gradient Gel Electrophoresis Profiles of 16S rRNA-Defined Populations Inhabiting a Hot Spring Microbial Mat Community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar] [PubMed]
- Harms, G.; Layton, A.C.; Dionisi, H.M.; Gregory, I.R.; Garrett, V.M.; Hawkins, S.A.; Robinson, K.G.; Sayler, G.S. Real-time PCR Quantification of Nitrifying Bacteria in a Municipal Wastewater Treatment Plant. Environ. Sci. Technol. 2003, 37, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S. The Biology Workbench—A Seamless Database and Analysis Environment for the Biologist. Proteins 1998, 32, 1–2. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Klappenbach, J.A.; Saxman, P.R.; Cole, J.R.; Schmidt, T.M. rrndb: The Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2008, 36, D25–D30. [Google Scholar] [CrossRef]
- American Public Health Association/American Water Works Association/Water Environment Federation. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association/American Water Works Association/Water Environment Federation: Washington, DC, USA, 1998. [Google Scholar]
- Ginige, M.P.; Carvalho, G.; Keller, J.; Blackall, L. Eco-Physiological Characterization of Fluorescence In Situ Hybridization Probe-Targeted Denitrifiers in Activated Sludge using Culture-Independent Methods. Lett. Appl. Microbiol. 2007, 44, 399–405. [Google Scholar] [CrossRef]
- Gumaelius, L.; Magnusson, G.; Pettersson, B.; Dalhammar, G. Comamonas denitrificans sp nov., an Efficient Denitrifying Bacterium Isolated from Activated Sludge. Int. J. Syst. Evolut. Microbiol. 2001, 51, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Su, J.J.; Liu, B.Y.; Liu, C.Y. Comparison of Aerobic Denitrification under High Oxygen Atmosphere by Thiosphaera pantotropha ATCC 35512 and Pseudomonas stutzeri SU2 Newly Isolated from the Activated Sludge of a Piggery Wastewater Treatment System. J. Appl. Microbiol. 2001, 90, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, M.; Rusanowska, P.; Jarzabek, J.; Nielsen, J.L. Community Dynamics of Denitrifying Bacteria in Full-Scale Wastewater Treatment Plants. Environ. Technol. 2016, 37, 2358–2367. [Google Scholar] [CrossRef]
- Zhang, T.; Shao, M.F.; Ye, L. 454 Pyrosequencing Reveals Bacterial Diversity of Activated Sludge from 14 Sewage Treatment Plants. ISME 2012, 6, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Marietou, A.; Richardson, D.; Cole, J.; Mohan, S. Nitrate Reduction by Desulfovibrio desulfuricans: A Periplasmic Nitrate Reductase System that Lacks NapB, but Includes a Unique Tetraheme c-Type Cytochrome, NapM. FEMS Microbiol. Lett. 2005, 248, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, D.A.; Gregory, L.G.; Carter, J.P.; Karakas-Sen, A.; Richardson, D.J.; Spiro, S. Detection of Genes for Periplasmic Nitrate Reductase in Nitrate Respiring Bacteria and in Community DNA. FEMS Microbiol. Lett. 1999, 177, 263–270. [Google Scholar] [CrossRef]
- Bru, D.; Sarr, A.; Philippot, L. Relative Abundances of Proteobacterial Membrane-Bound and Periplasmic Nitrate Reductases in Selected Environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef]
- Smith, C.J.; Nedwell, D.B.; Dong, L.F.; Osborn, A.M. Diversity and Abundance of Nitrate Reductase Genes (narG and napA) Nitrite Reductase Genes (nirS and nrfA), and their Transcripts in Estuarine Sediments. Appl. Environ. Microbiol. 2007, 73, 3612–3622. [Google Scholar] [CrossRef]
- Cooksley, C.; Jenks, P.J.; Green, A.; Cockayne, A.; Logan, R.P.H.; Hardie, K.R. napA Protects Helicobacter pylori from Oxidative Stress Damage, and its Production is Influenced by the Ferric Uptake Regulator. J. Med. Microbiol. 2003, 52, 461–469. [Google Scholar] [CrossRef]
- Kim, J.O.; Cho, K.H.; Ligaray, M.; Jang, H.M.; Kang, S.; Kim, Y.M. Monitoring Influential Environmental Conditions Affecting Communities of Denitrifying and Nitrifying Bacteria in a Combined Anoxic-Oxic Activated Sludge System. Int. Biodeterior. Biodegrad. 2015, 100, 1–6. [Google Scholar] [CrossRef]
- Do, H.; Lim, J.; Shin, S.G.; Wu, Y.J.; Ahn, J.H.; Hwang, S. Simultaneous Effect of Temperature, Cyanide and Ammonia-Oxidizing Bacteria Concentrations on Ammonia Oxidation. J. Ind. Microbiol. Biotechnol. 2008, 35, 1331–1338. [Google Scholar] [CrossRef] [PubMed]
- Gujer, W.; Henze, M.; Mino, T.; van Loosdrecht, M. Activated Sludge Model no. 3. Water Sci. Technol. 1999, 39, 183–193. [Google Scholar] [CrossRef]
- Kim, Y.M.; Cho, H.U.; Lee, D.S.; Park, D.; Park, J.M. Influence of Operational Parameters on Nitrogen Removal Efficiency and Microbial Communities in a Full-Scale Activated Sludge Process. Water Res. 2011, 45, 5785–5795. [Google Scholar] [CrossRef] [PubMed]
- Limpiyakorn, T.; Shinohara, Y.; Kurisu, F.; Yagi, O. Communities of Ammonia-Oxidizing Bacteria in Activated Sludge of Various Sewage Treatment Plants in Tokyo. FEMS Microbiol. Ecol. 2005, 54, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Juretschko, S.; Loy, A.; Lehner, A.; Wagner, M. The Microbial Community Composition of a Nitrifying-Denitrifying Activated Sludge from an Industrial Sewage Treatment Plant Analyzed by the Full-Cycle rRNA Approach. Syst. Appl. Microbiol. 2002, 25, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Zumft, W.G. Cell Biology and Molecular Basis of Denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar]
- Grady, C.P.L.; Daigger, G.T.; Lim, H.C. Biological Wastewater Treatment, 2nd ed.; Marcel Dekker: New York, NY, USA, 1999. [Google Scholar]
- Oh, J.; Silverstein, J. Oxygen Inhibition of Activated Sludge Denitrification. Water Res. 1999, 33, 1925–1937. [Google Scholar] [CrossRef]
- Kester, R.A.; De Boer, W.; Laanbroek, H.J. Production of NO and N2O by Pure Cultures of Nitrifying and Denitrifying Bacteria during Changes in Aeration. Appl. Environ. Microbiol. 1997, 63, 3872–3877. [Google Scholar]
- Hernandez, D.; Rowe, J.J. Oxygen Inhibition of Nitrate Uptake Is a General Regulatory Mechanism in Nitrate Respiration. J. Biol. Chem. 1988, 263, 7937–7939. [Google Scholar]
- Stensel, H.D.; Loehr, R.C.; Lawrence, A.W. Biological Kinetics of Suspended-Growth Denitrification. J. Water Pollut. Control Fed. 1973, 45, 249–261. [Google Scholar]
- Kornaros, M.; Zafiri, C.; Lyberatos, G. Kinetics of Denitrification by Pseudomonas denitrificans under Growth Conditions Limited by Carbon and/or Nitrate or Nitrite. Water Environ. Res. 1996, 68, 934–945. [Google Scholar] [CrossRef]
- Briones, A.; Raskin, L. Diversity and Dynamics of Microbial Communities in Engineered Environments and their Implications for Process Stability. Curr. Opin. Biotechnol. 2003, 14, 270–276. [Google Scholar] [CrossRef]
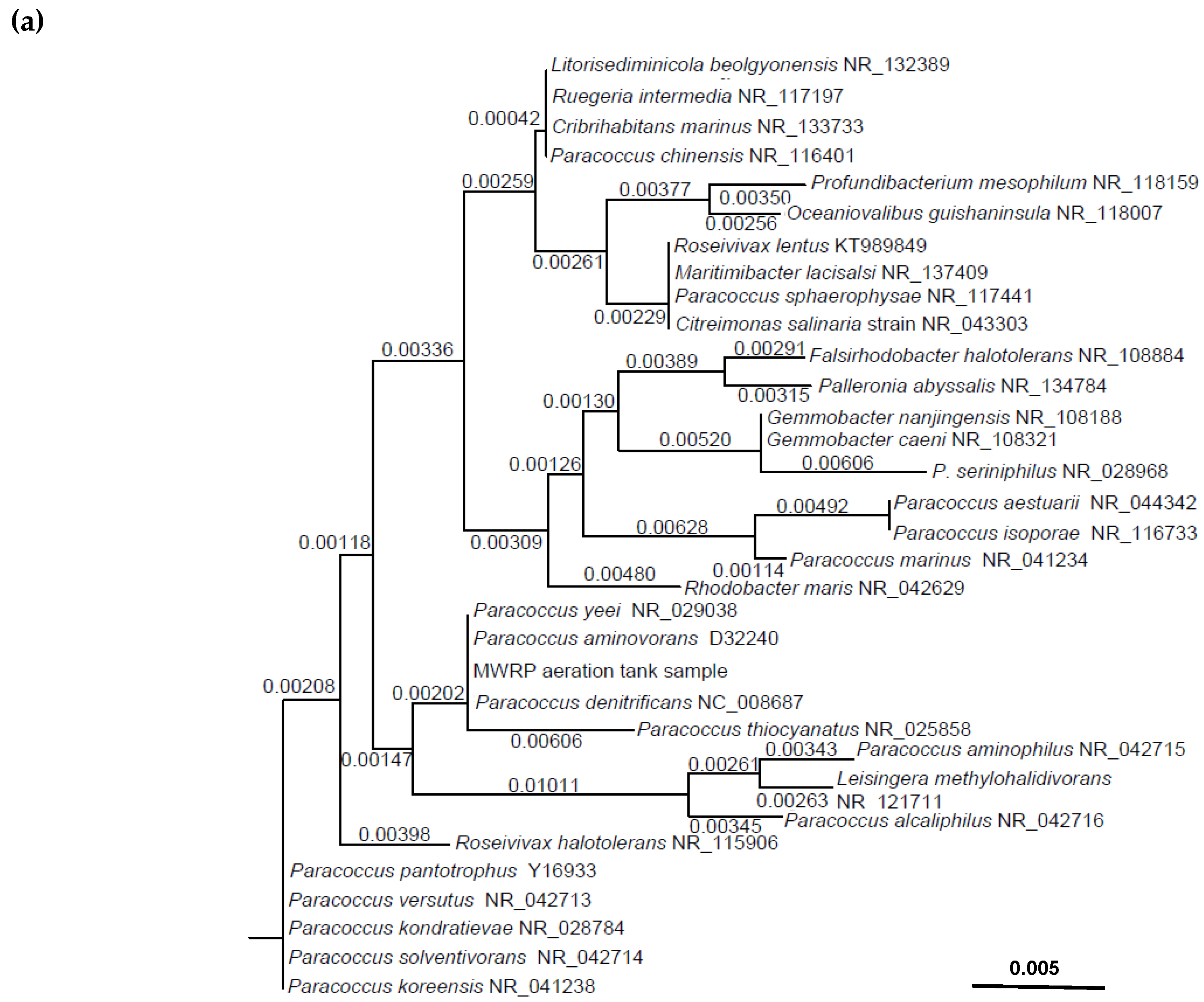
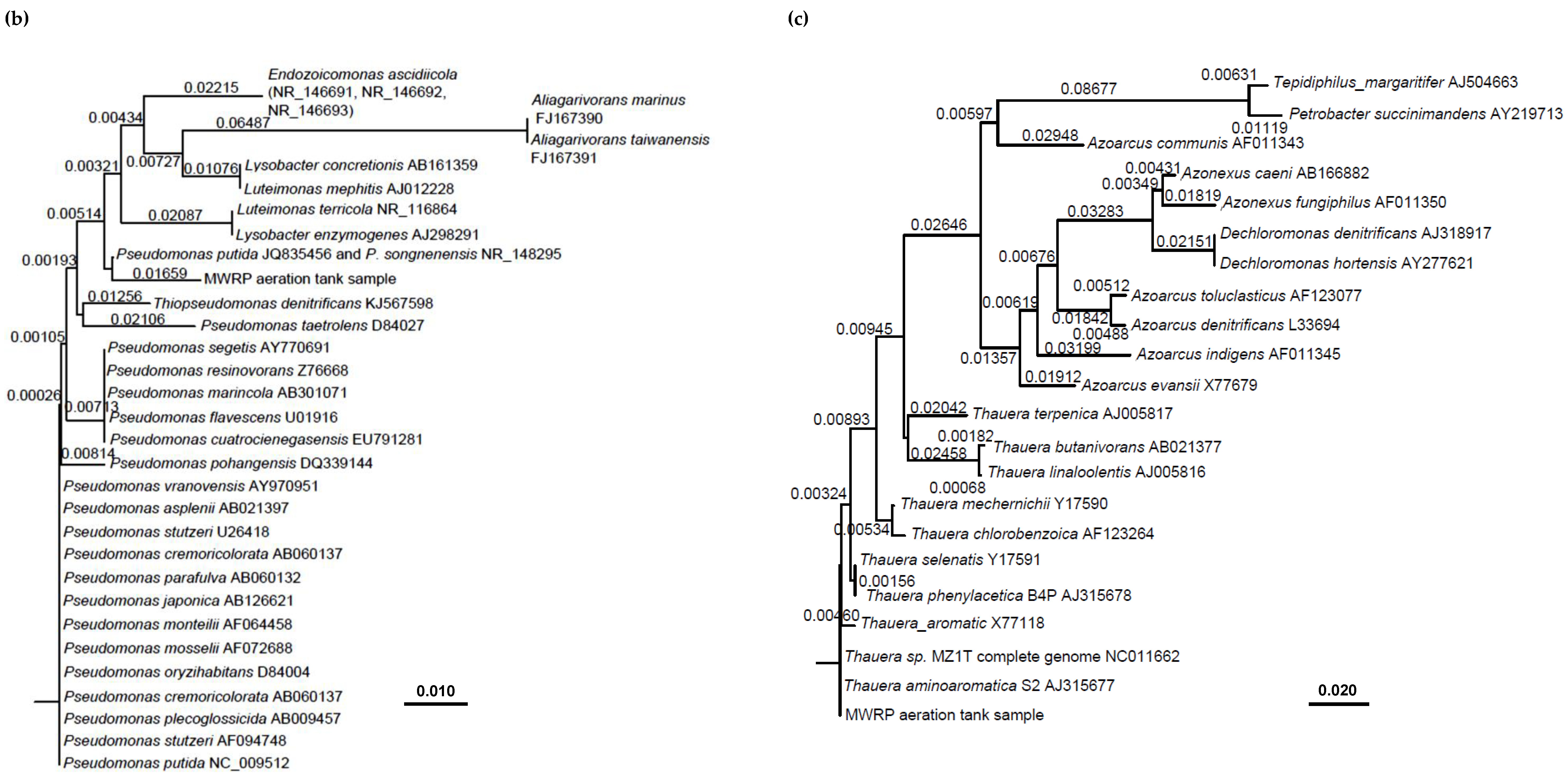
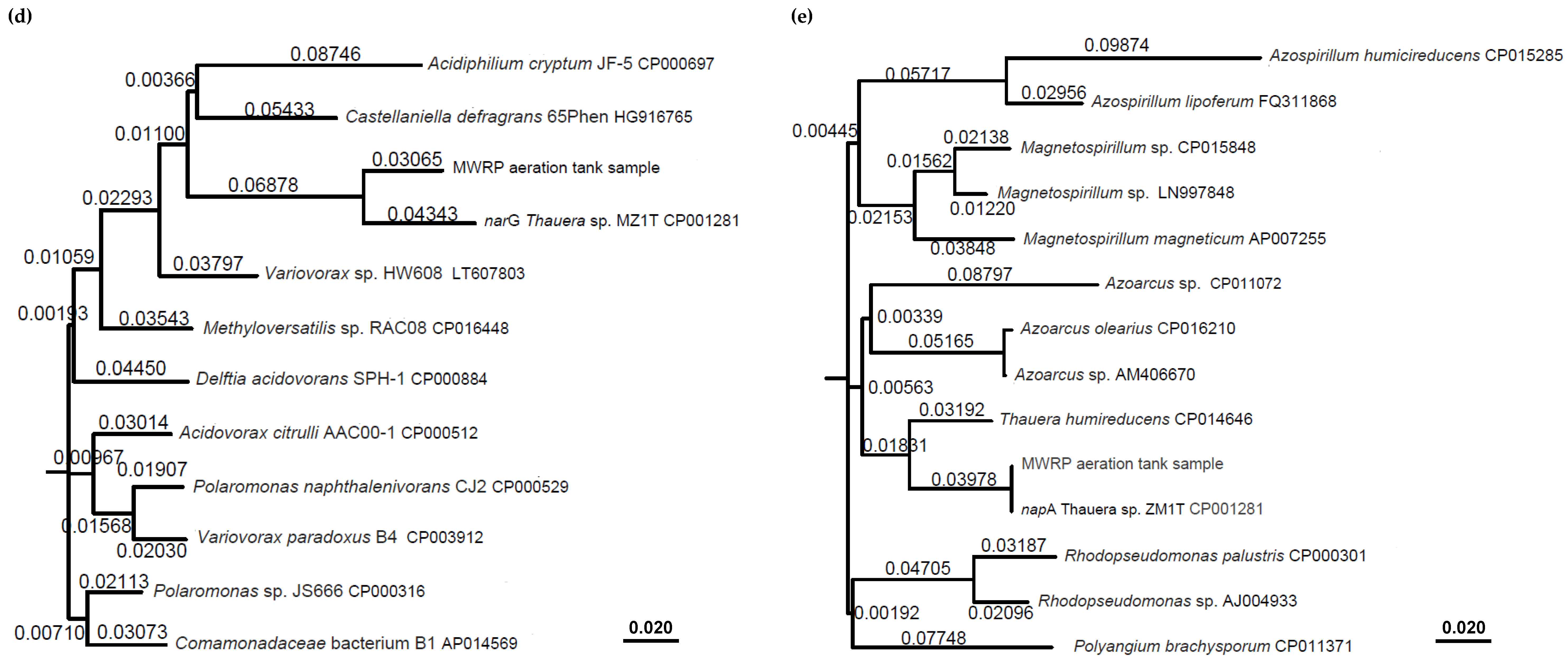


| Target Organisms/Genes | Primers and Probes | Sequence (5′–3′) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| Paracoccus-like spp. | 16S Para F | GGGCAGCATGCTGTTCGGTG | 60.0 | [18] |
| 16S Para R | GCGATGTCAAGGGTTGGTAA | |||
| 16S Para-Taq | ACACCTAACGGATTAAGCATTCCGCCTGGG | |||
| Pseudomonas spp. | 16S Pse F | GGATTGGTGCCTTCGGGAAC | 60.1 | [18] |
| 16S Pse R | ACGTGCTGGTAACTAAGGA | |||
| 16S Pse-Taq | CACAGGTGCTGCATGGCTGTCGTCAGCTCG | |||
| Thauera spp. | 16S Thau F | CGTGGTCTCTAACATAGGCC | 66.9 | [19] |
| 16S Thau R | CGGAGCAGCAATGCACTGAG | |||
| 16S Thau-Taq | TACCGGACTAAGAAGCACCGGCTAACTACGT | |||
| narG | NarG F | CAGGCGGCCGCGGATCATCGGG | 66 | [18] |
| NarG R | CAGCAGACCGACTACCCGCGC | |||
| napA | NapA F | CAGCCCATCGGCTCGTC | 66 | [18] |
| NapA R | AGAACGGCGAGTTCACG | |||
| Total bacteria | 1055f | ATGGCTGTCGTCAGCT | 50 | [20] |
| 1392r | ACGGGCGGTGTGTAC | [21] | ||
| 16STaq1115 | CAACGAGCGCAACCC | [22] |
| Groups | Copies/Cells | References |
|---|---|---|
| Total bacteria | 3.6 | [22] |
| Paracoccus spp. | 3 | [25] |
| Thauera spp. | 4 | [26] |
| Pseudomonas spp.-like | 4.6 | [25] |
| narG-like gene | 1 | [26] |
| napA gene | 1 | [26] |
| Parameters | Average | Standard Deviation | Pearson’s Correlation Coefficients (r) with TNeff |
|---|---|---|---|
| Temperature (°C) | 26.6 | 1.51 | −0.676 ** |
| pH | 7.1 | 0.11 | 0.369 * |
| DOanoxic (mg/L) | 0.23 | 0.09 | 0.361 * |
| DOaerobic (mg/L) | 2.82 | 0.23 | −0.230 |
| SRT (day) | 8.24 | 0.59 | −0.377 * |
| HRT (h) | 5.98 | 0.29 | −0.020 |
| MLSS (mg/L) | 2437 | 119 | −0.142 |
| Methanol (mg/L) | 9.06 | 0.92 | 0.181 |
| BODin (mg/L) | 146 | 13 | 0.320 * |
| CODin (mg/L) | 296 | 24 | 0.522 ** |
| Ammoniumin (mg-N/L) | 28.17 | 2.08 | 0.700 ** |
| TNin (mg-N/L) | 29.36 | 1.68 | 0.635 ** |
| BODeff (mg/L) | 7.66 | 2.22 | 0.660 ** |
| CODeff (mg/L) | 27.3 | 5.66 | 0.656 ** |
| Ammoniumeff (mg-N/L) | 0.18 | 0.08 | 0.355 * |
| TNeff (mg-N/L) | 9.12 | 0.81 | − |
| Denitrifier | Average (cells/L) | Standard Deviation (cells/L) | Pearson’s Correlation Coefficients with TNeff |
|---|---|---|---|
| Thauera | 2.86 × 1012 | 2.24 × 1012 | −0.793 ** |
| Paracoccus | 1.65 × 1010 | 7.92 × 109 | −0.543 ** |
| Pseudomonas-like | 3.61 × 109 | 2.65 × 109 | −0.404 ** |
| Total Bacteria | 1.80 × 1013 | 3.90 × 1012 | −0.352 * |
| narG-like | 2.10 × 1011 | 8.00 × 1010 | −0.663 ** |
| napA | 5.14 × 109 | 2.72 × 109 | −0.643 ** |
| Denitrifier | Thauera spp. | Paracoccus spp. | Pseudomonas spp.-like | Total Bacteria | narG-like | napA |
|---|---|---|---|---|---|---|
| Thauera spp. | 0.823 ** | 0.842 ** | 0.372 * | 0.891 ** | 0.890 ** | |
| Paracoccus spp. | 0.838 ** | 0.632 ** | 0.868 ** | 0.860 ** | ||
| Pseudomonas spp.-like | 0.426 ** | 0.815 ** | 0.845 ** | |||
| Total Bacteria | 0.426 ** | 0.424 ** | ||||
| narG-like | 0.852 ** |
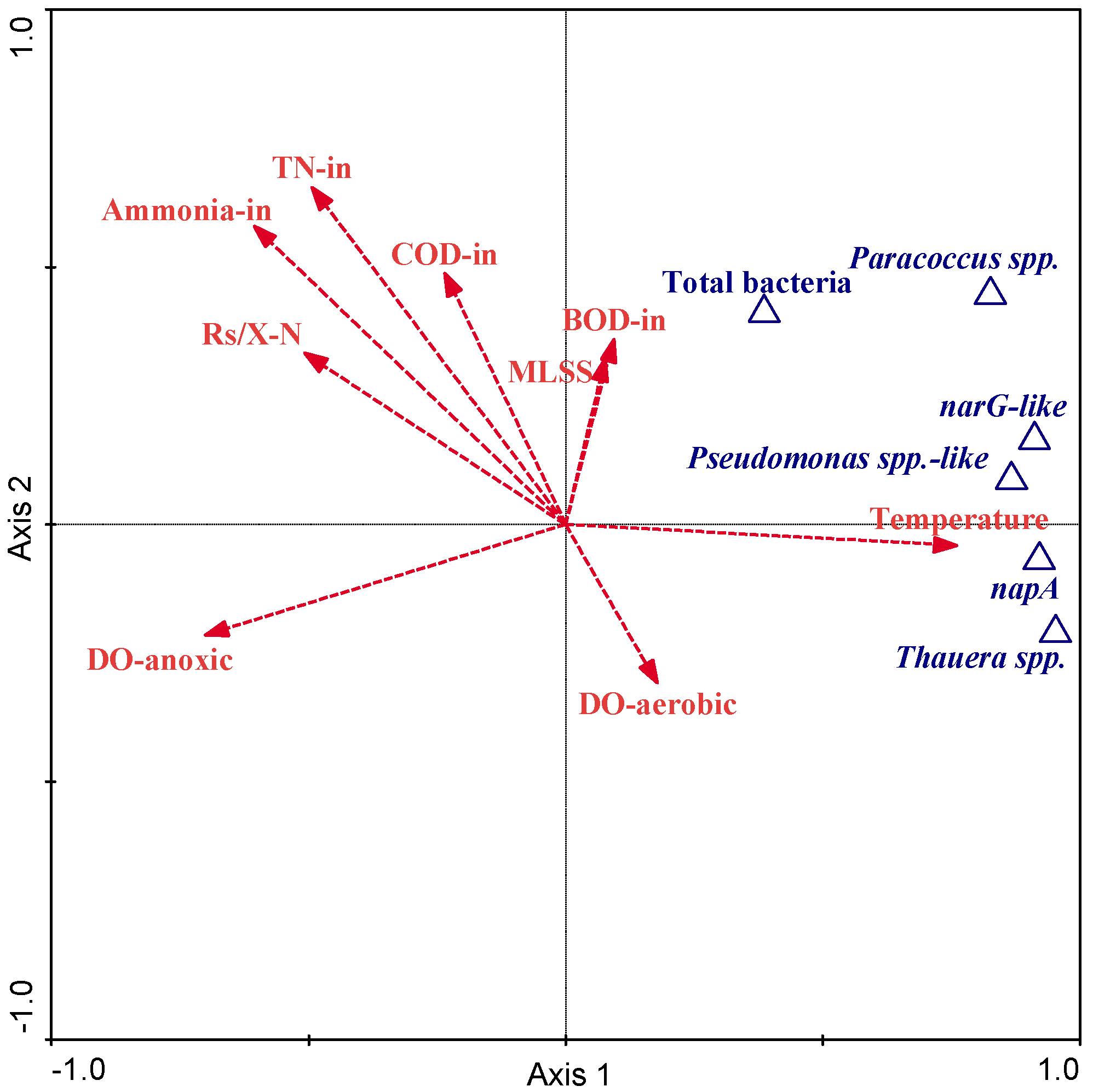
| Pearson’s Correlation Coefficients | RDA Forward Selection | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Thauera spp. | Paracoccus spp. | Pseudomonas spp.-like | Total bacteria | narG-like | napA | Ranking | p | F |
| Temperature | 0.739 ** | 0.705 ** | 0.462 ** | 0.201 | 0.669 ** | 0.796 ** | 1 | 0.002 | 34.25 |
| pH | −0.507 ** | −0.410 ** | −0.479 ** | −0.203 | −0.486 ** | −0.421 ** | 16 | 0.866 | 0.29 |
| DOanoxic | −0.600 ** | −0.674 ** | −0.700 ** | −0.190 | −0.675 ** | −0.636 ** | 3 | 0.002 | 10.77 |
| DOaerobic | 0.228 | 0.003 | 0.158 | −0.167 | 0.164 | 0.163 | 9 | 0.036 | 3.56 |
| SRT | 0.532 ** | 0.619 ** | 0.462 ** | 0.306 | 0.652 ** | 0.586 ** | 11 | 0.14 | 1.68 |
| HRT | 0.393 * | 0.190 | 0.551 ** | −0.207 | 0.270 | 0.413** | 10 | 0.056 | 3.24 |
| MLSS | 0.039 | 0.171 | −0.038 | 0.310 | 0.383 * | −0.009 | 7 | 0.008 | 5.35 |
| Methanol | 0.183 | 0.149 | 0.499 ** | −0.110 | 0.108 | 0.181 | 15 | 0.734 | 0.41 |
| BODin | 0.000 | 0.191 | 0.274 | 0.073 | 0.125 | −0.013 | 5 | 0.002 | 8.66 |
| CODin | −0.327 * | −0.035 | −0.009 | 0.066 | −0.117 | −0.359 * | 8 | 0.018 | 4.74 |
| Ammoniumin | −0.702 ** | −0.276 | −0.390 * | −0.014 | −0.439 ** | −0.646 ** | 2 | 0.004 | 13.48 |
| TNin | −0.609 ** | −0.160 | −0.259 | 0.075 | −0.331 * | −0.563 ** | 4 | 0.002 | 10.6 |
| F/M | −0.583 ** | −0.258 | −0.323 * | −0.014 | −0.509 ** | −0.596 ** | 12 | 0.232 | 1.38 |
| COD/TN | 0.205 | 0.155 | 0.324 * | 0.030 | 0.226 | 0.092 | 13 | 0.338 | 1 |
| BOD/COD | 0.442 ** | 0.328 * | 0.423 ** | −0.004 | 0.336 * | 0.473 ** | 14 | 0.526 | 0.69 |
| Rs /X-N | −0.582 ** | −0.285 | −0.207 | −0.182 | −0.596 ** | −0.508 ** | 6 | 0.002 | 7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Olson, B.H.; Asvapathanagul, P.; Wang, T.; Tsai, R.; Rosso, D. Molecular Biomarkers and Influential Factors of Denitrification in a Full-Scale Biological Nitrogen Removal Plant. Microorganisms 2020, 8, 11. https://doi.org/10.3390/microorganisms8010011
Fang H, Olson BH, Asvapathanagul P, Wang T, Tsai R, Rosso D. Molecular Biomarkers and Influential Factors of Denitrification in a Full-Scale Biological Nitrogen Removal Plant. Microorganisms. 2020; 8(1):11. https://doi.org/10.3390/microorganisms8010011
Chicago/Turabian StyleFang, Hua, Betty H Olson, Pitiporn Asvapathanagul, Tongzhou Wang, Raymond Tsai, and Diego Rosso. 2020. "Molecular Biomarkers and Influential Factors of Denitrification in a Full-Scale Biological Nitrogen Removal Plant" Microorganisms 8, no. 1: 11. https://doi.org/10.3390/microorganisms8010011
APA StyleFang, H., Olson, B. H., Asvapathanagul, P., Wang, T., Tsai, R., & Rosso, D. (2020). Molecular Biomarkers and Influential Factors of Denitrification in a Full-Scale Biological Nitrogen Removal Plant. Microorganisms, 8(1), 11. https://doi.org/10.3390/microorganisms8010011






