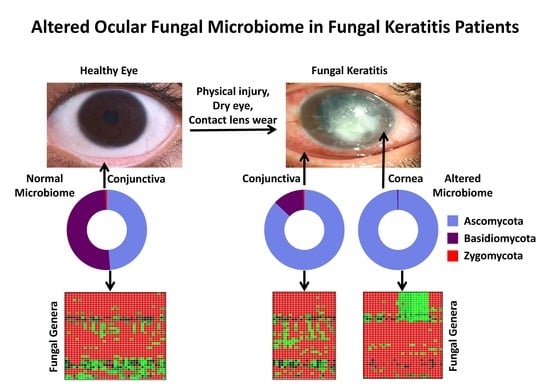
Alterations in the Ocular Surface Fungal Microbiome in Fungal Keratitis Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Subjects
2.2. Sample Collection
2.3. Culturing of Fungi
2.4. DNA Extraction
2.5. PCR Amplification, Illumina Library Preparation, and Amplicon Sequencing
2.6. Taxonomy Assignment of Sequenced Reads
2.7. Diversity Analyses of the Microbiomes
2.8. Identification of Differentially Abundant Genera
2.9. Principal Coordinate Analysis of the Microbiomes of Conjunctival Swabs and Corneal Scrapings of Fungal Keratitis Patients Who Have Taken Antifungal Medication
2.10. Interaction Networks between Fungal Genera in the Microbiomes
2.11. Correlation of Fungal Genera in HC-SW, FK-SW, and FK-CR
3. Results
3.1. Sample Details
3.2. Detection of Fungi by Culturable Approach
3.3. NGS Analysis of the Fungal Ocular Microbiomes
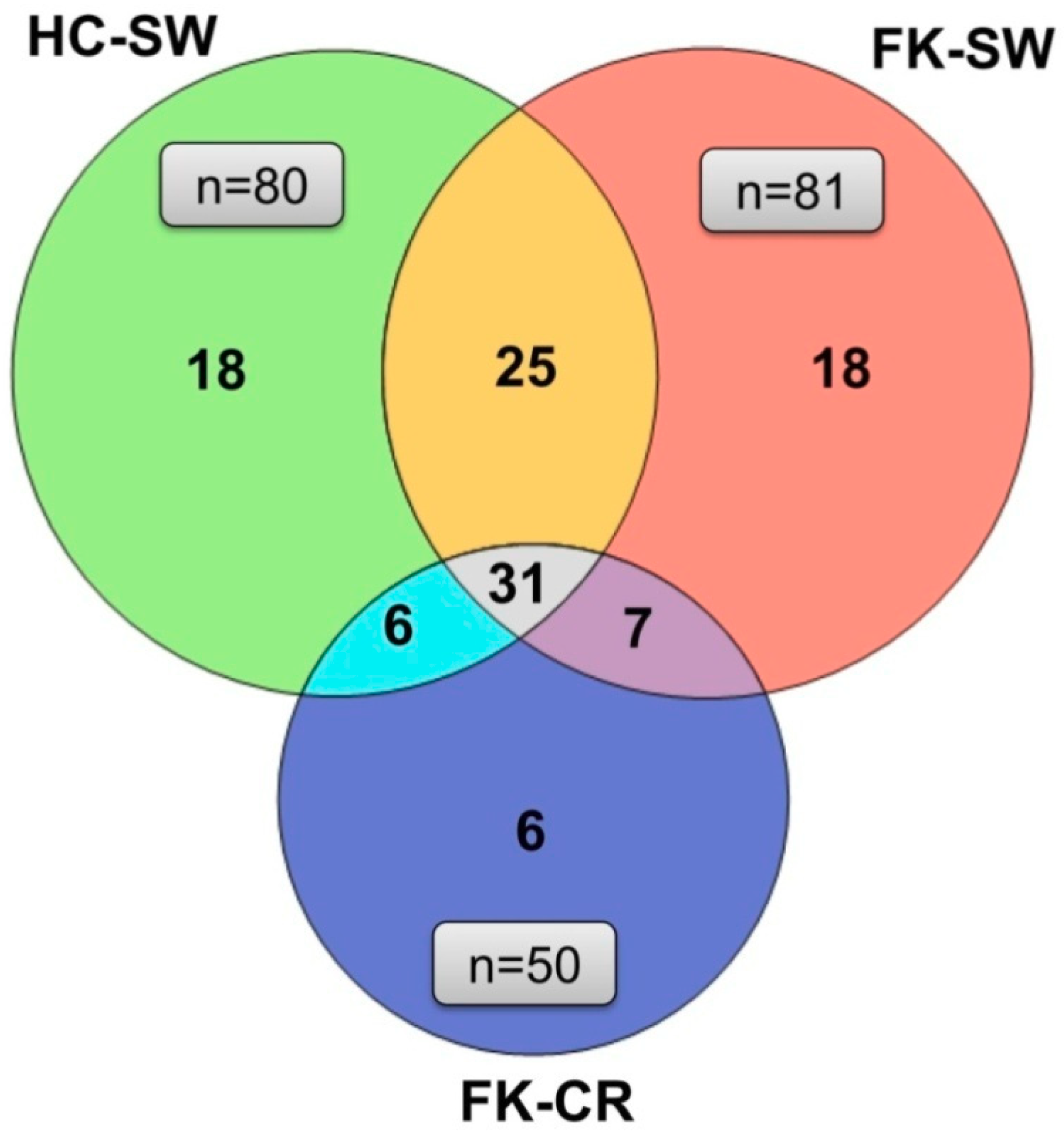
3.4. OTU Analysis
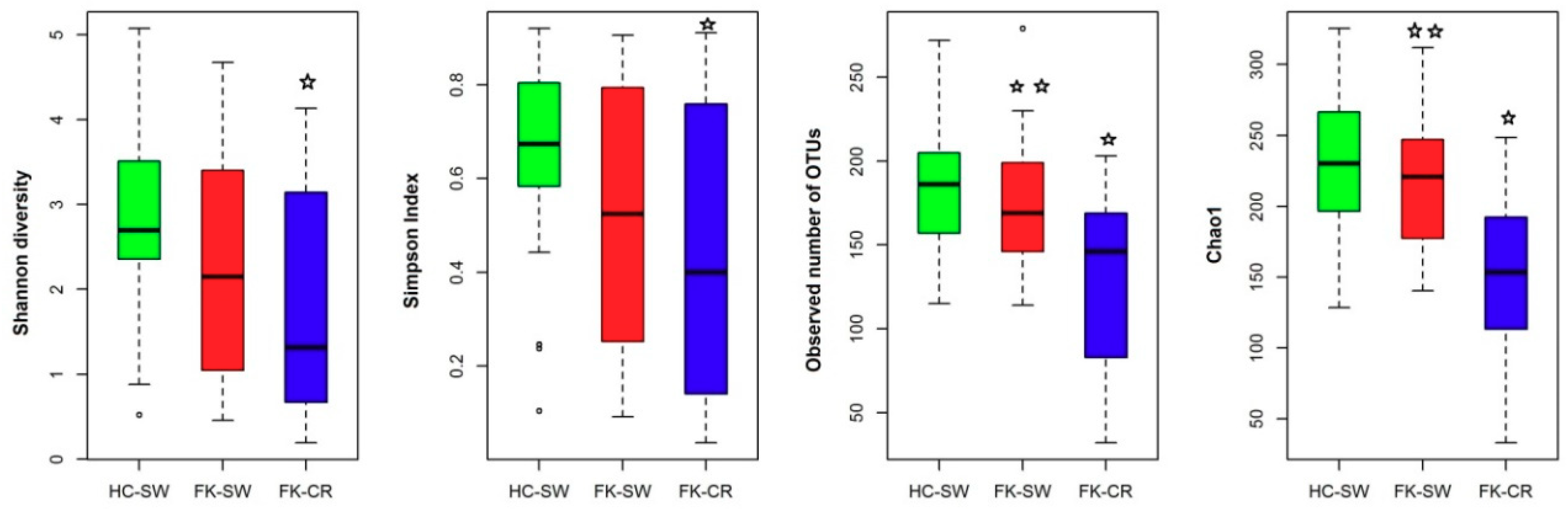
3.5. Alpha Diversity Indices of the Ocular Fungal Microbiomes
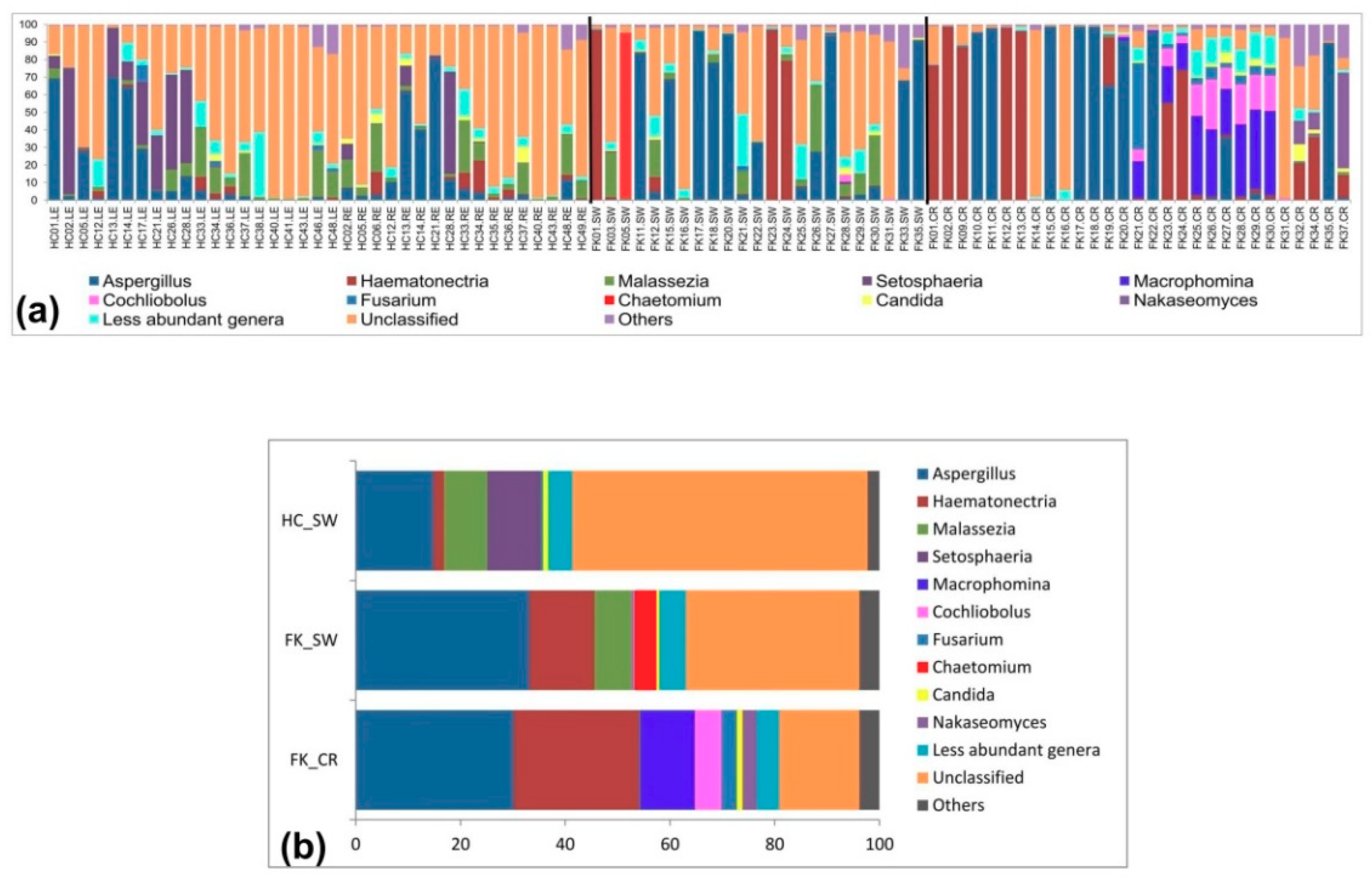
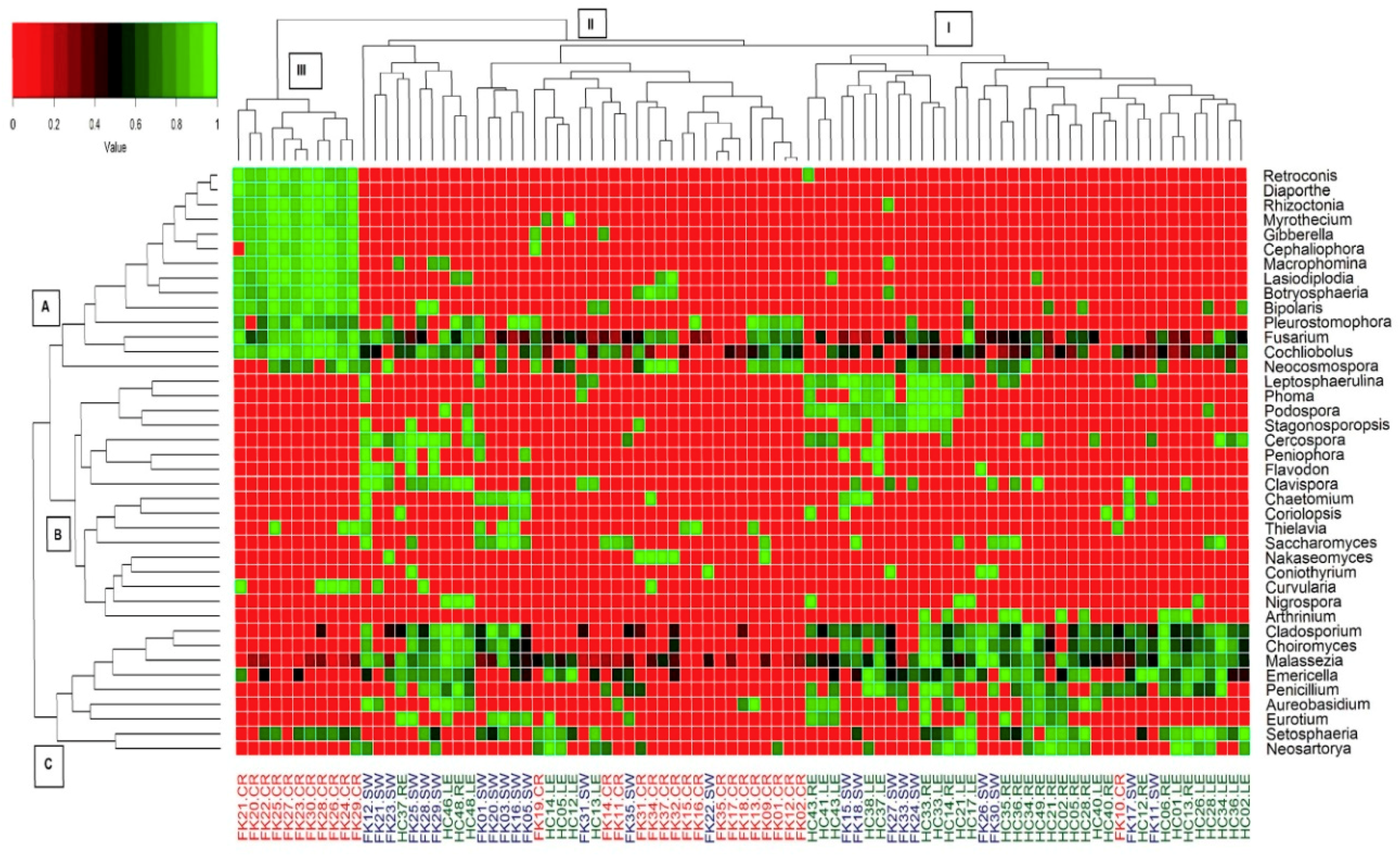
3.6. Fungal Community Composition of the Ocular Microbiomes
3.7. Principal Coordinate Analysis of the Microbiomes of Conjunctival Swabs and Corneal Scrapings of Fungal Keratitis Patients Who Have Taken Antifungal Medication
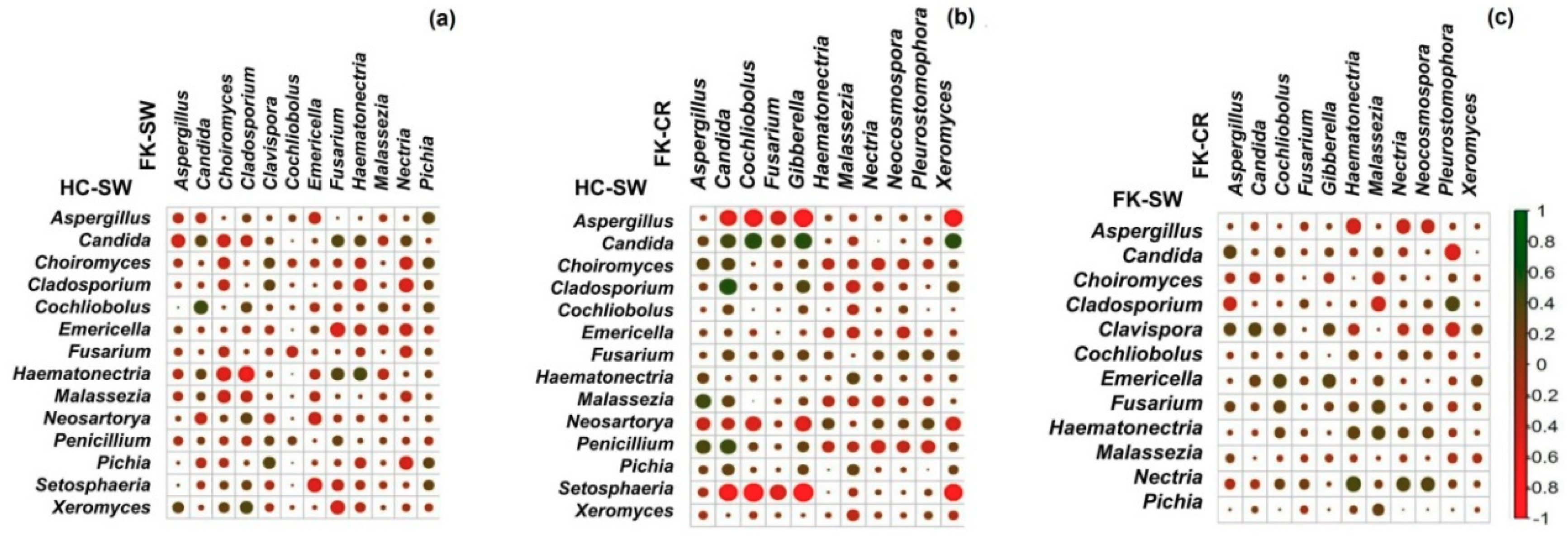
3.8. Interactions between Fungal Groups Inhabiting the Ocular Surface of Healthy Subjects and FK Patients
3.9. Correlation of Fungal Genera in HC-SW, FK-SW, and FK-CR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Tlaskalova-Hogenova, H.; Stepankova, R.; Kozakova, H.; Hudcovic, T.; Vannucci, L.; Tuckova, L.; Rossmann, P.; Hrncir, T.; Kverka, M.; Zakostelska, Z.; et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 2011, 8, 110–120. [Google Scholar] [CrossRef]
- Zackular, J.P.; Baxter, N.T.; Iverson, K.D.; Sadler, W.D.; Petrosino, J.F.; Chen, G.Y.; Schloss, P.D. The gut microbiome modulates colon tumorigenesis. MBio. 2013. [Google Scholar] [CrossRef]
- Malan-Muller, S.; Valles-Colomer, M.; Raes, J.; Lowry, C.A.; Seedat, S.; Hemmings, S.M.J. The Gut Microbiome and Mental Health: Implications for Anxiety- and Trauma-Related Disorders. OMICS. 2018, 22, 90–107. [Google Scholar] [CrossRef]
- Shivaji, S. We are not alone: A case for the human microbiome in extra intestinal diseases. Gut Pathog. 2017, 9, 13. [Google Scholar] [CrossRef][Green Version]
- Horai, R.; Zarate-Blades, C.R.; Dillenburg-Pilla, P.; Chen, J.; Kielczewski, J.L.; Silver, P.B.; Jittayasothorn, Y.; Chan, C.C.; Yamane, H.; Honda, K.; et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity 2015, 43, 343–353. [Google Scholar] [CrossRef]
- Huang, X.; Ye, Z.; Cao, Q.; Su, G.; Wang, Q.; Deng, J.; Zhou, C.; Kijlstra, A.; Yang, P. Gut Microbiota Composition and Fecal Metabolic Phenotype in Patients With Acute Anterior Uveitis. Invest. Ophthalmol. Vis. Sci. 2018, 59, 1523–1531. [Google Scholar] [CrossRef]
- Jayasudha, R.; Kalyana Chakravarthy, S.; Sai Prashanthi, G.; Sharma, S.; Tyagi, M.; Shivaji, S. Implicating Dysbiosis of the Gut Fungal Microbiome in Uveitis, an Inflammatory Disease of the Eye. Invest. Ophthalmol. Vis. Sci. 2019, 60, 1384–1393. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Sai Prashanthi, G.; Ali, M.H.; Sharma, S.; Tyagi, M.; Shivaji, S. Dysbiosis in the Gut Bacterial Microbiome of Patients with Uveitis, an Inflammatory Disease of the Eye. Indian J. Microbiol. 2018, 58, 457–469. [Google Scholar] [CrossRef]
- Shimizu, J.; Kubota, T.; Takada, E.; Takai, K.; Fujiwara, N.; Arimitsu, N.; Ueda, Y.; Wakisaka, S.; Suzuki, T.; Suzuki, N. Bifidobacteria Abundance-Featured Gut Microbiota Compositional Change in Patients with Behcet′s Disease. PLoS ONE 2016, 11, e0153746. [Google Scholar] [CrossRef]
- de Paiva, C.S.; Jones, D.B.; Stern, M.E.; Bian, F.; Moore, Q.L.; Corbiere, S.; Streckfus, C.F.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci. Rep. 2016, 1, 23561. [Google Scholar] [CrossRef]
- Rowan, S.; Jiang, S.; Korem, T.; Szymanski, J.; Chang, M.L.; Szelog, J.; Cassalman, C.; Dasuri, K.; McGuire, C.; Nagai, R.; et al. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef]
- Zinkernagel, M.S.; Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; Leichtle, A.B.; Largiadèr, C.R.; Fiedler, G.M.; Wolf, S. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci. Rep. 2017, 7, 40826. [Google Scholar] [CrossRef]
- Jayasudha, R.; Chakravarthy, S.K.; Prashanthi, G.S.; Sharma, S.; Garg, P.; Murthy, S.I.; Shivaji, S. Alterations in gut bacterial and fungal microbiomes are associated with bacterial Keratitis, an inflammatory disease of the human eye. J. Biosci. 2018, 43, 835–856. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Ranjith, K.; Dutta, A.; Pinna, N.K.; Mande, S.S.; Sharma, S.; Garg, P.; Murthy, S.I.; Shivaji, S. Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS ONE 2018, 13, e0199640. [Google Scholar] [CrossRef]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal Stability and Composition of the Ocular Surface Microbiome. Sci. Rep. 2017, 7, 9880. [Google Scholar] [CrossRef]
- Segal, E.; Romano, A.; Eylan, E.; Stein, R. Fungal flora of the normal conjunctival sac. Mycoses 1977, 20, 9–14. [Google Scholar] [CrossRef]
- Wu, T.G.; Mitchell, B.M.; Carothers, T.S.; Coats, D.K.; Brady-McCreery, K.M.; Paysse, E.A.; Wilhelmus, K.R. Molecular analysis of the pediatric ocular surface for fungi. Curr. Eye Res. 2003, 26, 33–36. [Google Scholar] [CrossRef]
- Shivaji, S.; Jayasudha, R.; Sai Prashanthi, G.; Kalyana Chakravarthy, S.; Sharma, S. The Human Ocular Surface Fungal Microbiome. Invest. Ophthalmol. Vis. Sci. 2019, 60, 451–459. [Google Scholar] [CrossRef]
- Dadaci, Z.; Kilinc, F.; Ozer, T.T.; Sahin, G.O.; Acir, N.O.; Borazan, M. Periodic acid-Schiff staining demonstrates fungi in chronic anterior blepharitis. Eye (Lond) 2015. [Google Scholar] [CrossRef]
- Zago, V.V.; Castro, M.A.; Tackman, R.N. Support of the Laboratory in the Diagnosis of Fungal Ocular Infections. Int. J. Inflamm. 2012. [Google Scholar] [CrossRef]
- Henry, C.R.; Flynn, H.W.; Miller, D.; Forster, R.K.; Alfonso, E.C. Infectious Keratitis Progressing to Endophthalmitis: A 15-Year-Study of Microbiology, Associated Factors, and Clinical Outcomes. Ophthalmology 2012, 119, 2443–2449. [Google Scholar] [CrossRef]
- Teweldemedhin, M.; Gebreyesus, H.; Atsbaha, A.H.; Asgedom, S.W.; Saravanan, M. Bacterial profile of ocular infections: A systematic review. BMC Ophthalmol. 2017, 17, 212. [Google Scholar] [CrossRef]
- Gonzales, C.A.; Srinivasan, M.; Whitcher, J.P.; Smolin, G. Incidence of corneal ulceration in Madurai district, South India. Ophthalmic Epidemiol. 1996, 3, 159–166. [Google Scholar] [CrossRef]
- Gupta, N.; Tandon, R.; Gupta, S.K.; Sreenivas, V.; Vashist, P. Burden of Corneal Blindness in India. Indian J. Community Med. 2013, 38, 198–206. [Google Scholar] [CrossRef]
- Prajna, V.; Prajna, L.; Muthiah, S. Fungal keratitis: The Aravind experience. Indian J. Ophthalmol. 2017, 65, 912–919. [Google Scholar] [CrossRef]
- Kunimoto, D.Y.; Sharma, S.; Garg, P.; Gopinathan, U.; Miller, D.; Rao, G.N. Corneal ulceration in the elderly in Hyderabad, south India. Br. J. Ophthalmol. 2000, 84, 54–59. [Google Scholar] [CrossRef]
- Rytas, V.; Hibbett, D.S.; John, S.; Hopple, J. Phylogenetic implications of generic concepts in fungal taxonomy: The impact of molecular systematic studies. Mycol. Helv. 1994, 6, 73–91. [Google Scholar]
- Dehingia, M.; Thangjam devi, K.; Talukdar, N.C.; Talukdar, R.; Reddy, N.; Mande, S.S.; Deka, M.; Khan, M.R. Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 2015, 5, 18563. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from Microbial Communities. Curr. Protoc. Bioinform. 2011. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Env. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Res. 2016, 5, 1519. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Abelson, M.B.; Lane, K.; Slocum, C. The Secrets of Ocular Microbiomes. 2015. Available online: http://www. reviewofophthalmology com/content/t/ocular_disease/c/55178/ (accessed on 5 December 2016).
- St Leger, A.J.; Desai, J.V.; Drummond, R.A.; Kugadas, A.; Almaghrabi, F.; Silver, P.; Raychaudhuri, K.; Gadjeva, M.; Iwakura, Y.; Lionakis, M.S.; et al. An Ocular Commensal Protects against Corneal Infection by Driving an Interleukin-17 Response from Mucosal γδ T Cells. Immunity 2017, 47, 148–158.e145. [Google Scholar] [CrossRef]
- Ge, C.; Wei, C.; Yang, B.-X.; Cheng, J.; Huang, Y.-S. Conjunctival microbiome changes associated with fungal keratitis: Metagenomic analysis. Int. J. Ophthalmol. 2019, 12, 194–200. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, B.; Li, W. Defining the normal core microbiome of conjunctival microbial communities. Clin. Microbiol. Infect. 2016, 22, 643.e7–643.e12. [Google Scholar] [CrossRef]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of bacteria at healthy human conjunctiva. Invest. Ophthalmol. Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef]
- Graham, J.E.; Moore, J.E.; Jiru, X.; Goodall, E.A.; Dooley, J.S.; Hayes, V.E.; Dartt, D.A.; Downes, C.S.; Moore, T.C. Ocular pathogen or commensal: A PCR-based study of surface bacterial flora in normal and dry eyes. Invest. Ophthalmol. Vis. Sci. 2007, 48, 5616–5623. [Google Scholar] [CrossRef]
- Karsten, E.; Watson, S.L.; Foster, L.J.R. Diversity of microbial species implicated in keratitis: A review. Open Ophthalmol. J. 2012, 6, 110–124. [Google Scholar] [CrossRef]
- Rosenbaum, J.T.; Lin, P.; Asquith, M. Does the Microbiome Cause B27-related Acute Anterior Uveitis? Ocul. Immunol. Inflamm. 2016, 24, 440–444. [Google Scholar] [CrossRef]
- Nakamura, Y.K.; Metea, C.; Karstens, L.; Asquith, M.; Gruner, H.; Moscibrocki, C.; Lee, I.; Brislawn, C.J.; Jansson, J.K.; Rosenbaum, J.T.; et al. Gut microbial alterations associated with protection from autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 2016, 57, 3747–3758. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project: Exploring the microbial part of ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Zhou, Y.; Holland, M.J.; Makalo, P.; Joof, H.; Roberts, C.H.; Mabey, D.C.; Bailey, R.L.; Burton, M.J.; Weinstock, G.M.; Burr, S.E. The conjunctival microbiome in health and trachomatous disease: A case control study. Genome Med. 2014, 6, 99. [Google Scholar] [CrossRef]
- Lee, S.H.; Oh, D.H.; Jung, J.Y.; Kim, J.C.; Jeon, C.O. Comparative Ocular Microbial Communities in Humans with and without Blepharitis. Invest. Ophthalmol. Vis. Sci. 2012, 53, 5585–5593. [Google Scholar] [CrossRef]
- Shin, H.; Price, K.; Albert, L.; Dodick, J.; Park, L.; Dominguez-Bello, M.G. Changes in the Eye Microbiota Associated with Contact Lens Wearing. MBio. 2016, 7, e00198-00116. [Google Scholar] [CrossRef]
- Revankar, S.G.; Sutton, D.A. Melanized fungi in human disease. Clin. Microbiol. Rev. 2010, 23, 884–928. [Google Scholar] [CrossRef]
- Srinivasan, S.; Hoffman, N.G.; Morgan, M.T.; Matsen, F.A.; Fiedler, T.L.; Hall, R.W.; Ross, F.J.; McCoy, C.O.; Bumgarner, R.; Marrazzo, J.M.; et al. Bacterial Communities in Women with Bacterial Vaginosis: High Resolution Phylogenetic Analyses Reveal Relationships of Microbiota to Clinical Criteria. PLoS ONE 2012, 7, e37818. [Google Scholar] [CrossRef]



| HQ Reads | HC-SW | FK-SW | FK-CR |
|---|---|---|---|
| (n = 37) | (n = 23) | (n = 29) | |
| Total | 15,465,381 | 10,756,288 | 10,909,499 |
| Maximum | 1,371,325 | 883,303 | 980,760 |
| Minimum | 88,897 | 61,695 | 153,623 |
| Average | 417,983.3 | 467,664.7 | 376,189.6 |
| HC-SW | FK-SW | FK-CR | p-Value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sl. No. | Phylum | Mean | Range | Present out of 37 Samples | Mean | Range | Present out of 23 Samples | Mean | Range | Present out of 29 Samples | HC-SW vs. FK-SW vs. FK-CR | HC-SW vs. FK-SW | HC–SW vs. FK-CR | FK-SW vs. FK-CR |
| 1 | Ascomycota | 35.66 | 0.5–98.78 | 37 | 69.99 | 9.86–98.23 | 23 | 88.36 | 2.22–99.24 | 29 | 0.001 | 0.001 | 0.001 | 0.006 |
| 2 | Basidiomycota | 37.05 | 0.34–95.75 | 37 | 10.12 | 0.01–46.11 | 23 | 0.35 | 0–2.15 | 23 | 0.001 | 0.001 | 0.001 | 0.001 |
| 3 | Zygomycota | 0.05 | 0–0.98 | 2 | 0 | 0–0.07 | 1 | 0 | 0–0 | 0 | 0.461 | 0.845 | 0.215 | 0.278 |
| 4 | Unclassified | 24.96 | 0.54–90.22 | 37 | 16.07 | 0.09–60.17 | 23 | 7.47 | 0–94.56 | 29 | 0.001 | 0.012 | 0.001 | 0.006 |
| 5 | Others * | 2.28 | 0.29–16.88 | 37 | 3.82 | 0.71–24.93 | 23 | 3.82 | 0.38–23.84 | 29 | NA | NA | NA | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sai Prashanthi, G.; Jayasudha, R.; Chakravarthy, S.K.; Padakandla, S.R.; SaiAbhilash, C.R.; Sharma, S.; Bagga, B.; Murthy, S.I.; Garg, P.; Shivaji, S. Alterations in the Ocular Surface Fungal Microbiome in Fungal Keratitis Patients. Microorganisms 2019, 7, 309. https://doi.org/10.3390/microorganisms7090309
Sai Prashanthi G, Jayasudha R, Chakravarthy SK, Padakandla SR, SaiAbhilash CR, Sharma S, Bagga B, Murthy SI, Garg P, Shivaji S. Alterations in the Ocular Surface Fungal Microbiome in Fungal Keratitis Patients. Microorganisms. 2019; 7(9):309. https://doi.org/10.3390/microorganisms7090309
Chicago/Turabian StyleSai Prashanthi, Gumpili, Rajagopalaboopathi Jayasudha, Sama Kalyana Chakravarthy, Shalem Raj Padakandla, Chinthala Reddy SaiAbhilash, Savitri Sharma, Bhupesh Bagga, Somasheila I. Murthy, Prashant Garg, and Sisinthy Shivaji. 2019. "Alterations in the Ocular Surface Fungal Microbiome in Fungal Keratitis Patients" Microorganisms 7, no. 9: 309. https://doi.org/10.3390/microorganisms7090309
APA StyleSai Prashanthi, G., Jayasudha, R., Chakravarthy, S. K., Padakandla, S. R., SaiAbhilash, C. R., Sharma, S., Bagga, B., Murthy, S. I., Garg, P., & Shivaji, S. (2019). Alterations in the Ocular Surface Fungal Microbiome in Fungal Keratitis Patients. Microorganisms, 7(9), 309. https://doi.org/10.3390/microorganisms7090309