Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Probiotic Products
2.2. Selective Culturing
2.3. Viability and Quantification Assessment
2.4. DNA Isolation
2.5. PCR for Selected Species
2.6. DNA Library Preparation and Sequencing
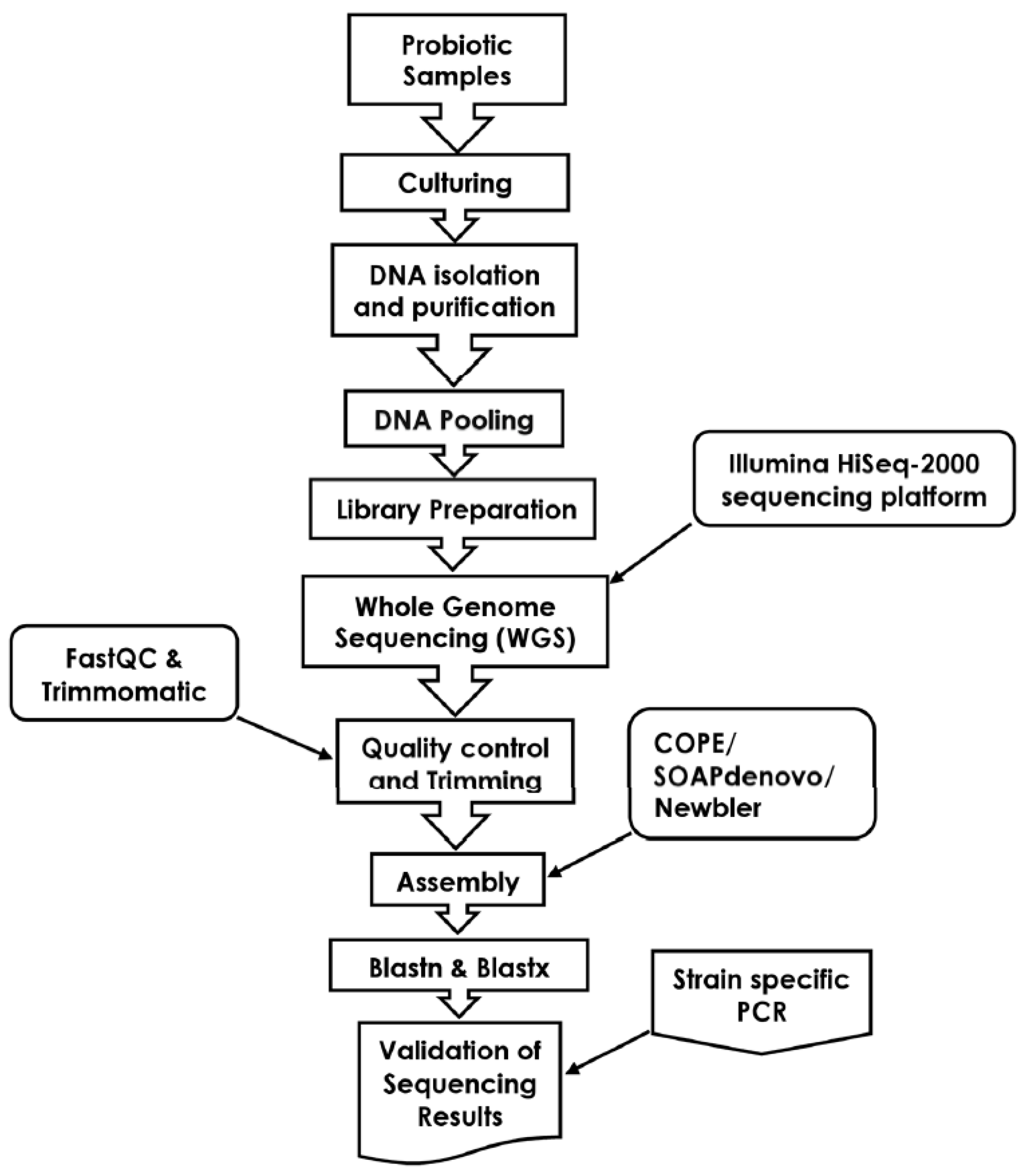
2.7. Sequence Analysis
2.8. Validation and Confirmation of Sequencing Results
2.9. Validation of Nonculturable Bacterial Contents by PCR
2.10. Availability of Data
3. Results
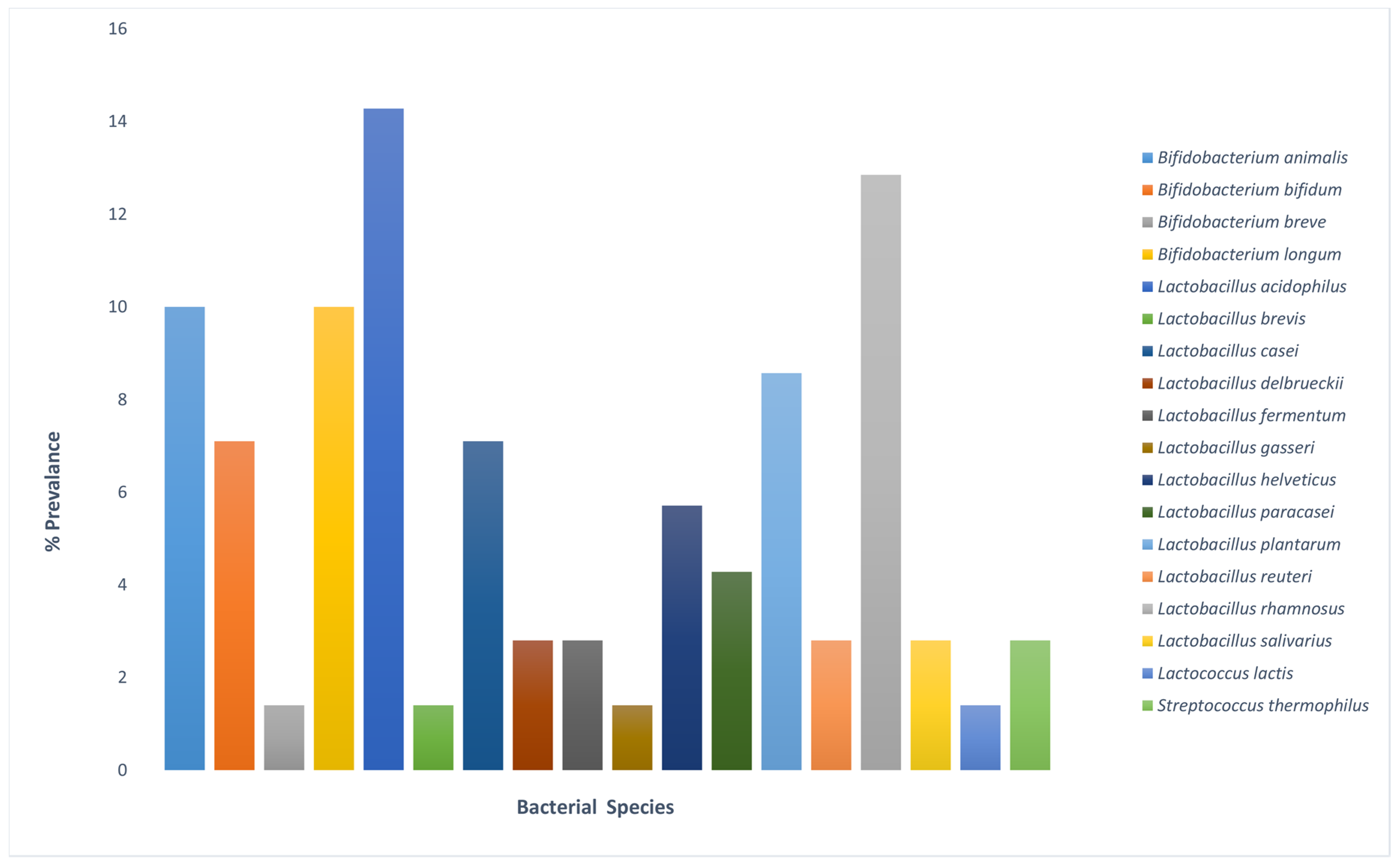
3.1. Microbial Viability and Quantification
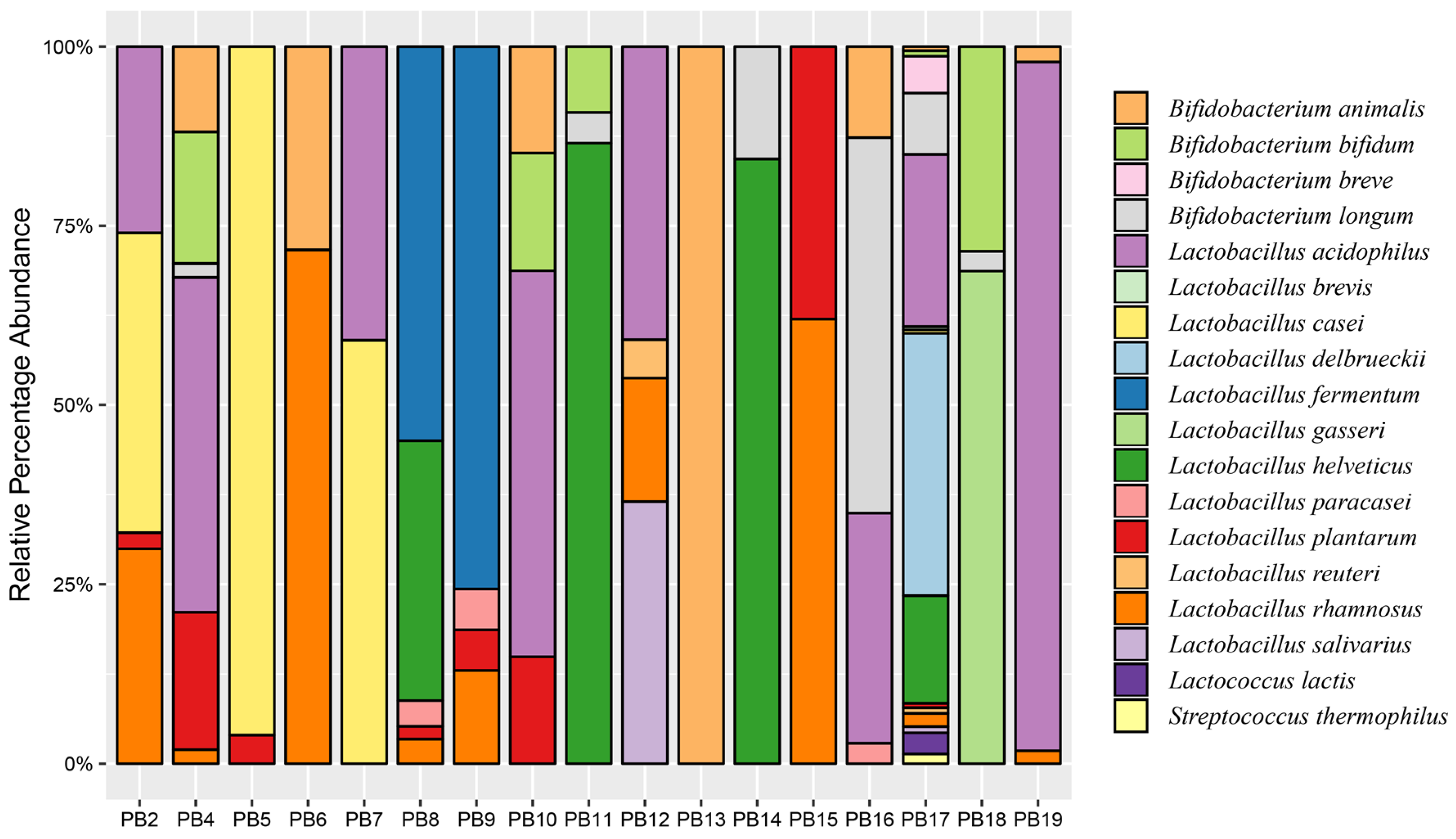
3.2. Sequence Analysis of Probiotic Products
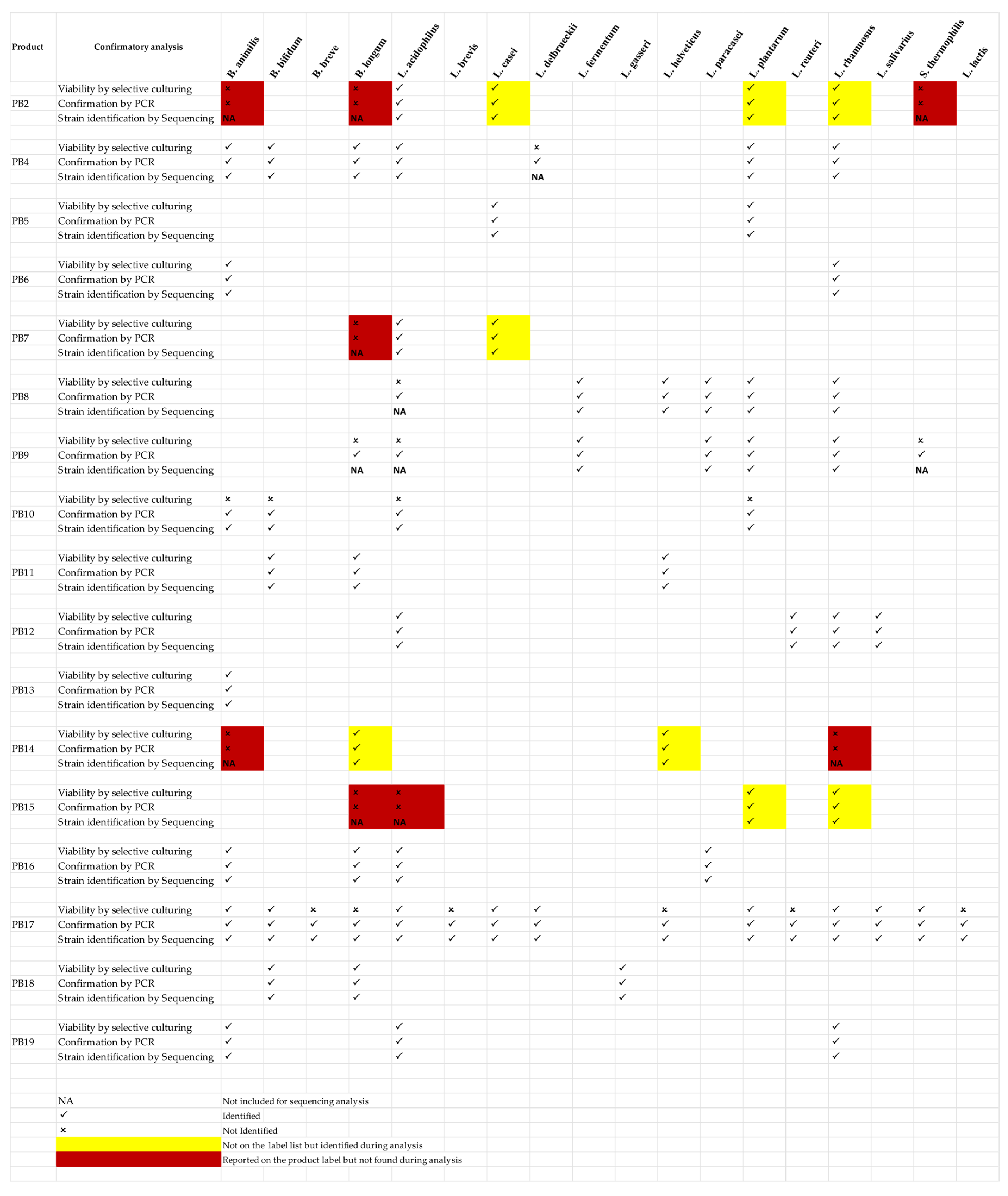
3.3. Confirmation of Sequence Analysis
3.4. Validation of Nonculturable Probiotic Bacterial Contents
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506. [Google Scholar] [CrossRef] [PubMed]
- Reid, G. The scientific basis for probiotic strains ofLactobacillus. Appl. Environ. Microbiol. 1999, 65, 3763–3766. [Google Scholar]
- Vankerckhoven, V.; Huys, G.; Vancanneyt, M.; Vael, C.; Klare, I.; Romond, M.-B.; Entenza, J.M.; Moreillon, P.; Wind, R.D.; Knol, J. Biosafety assessment of probiotics used for human consumption: Recommendations from the EU-PROSAFE project. Trends Food Sci. Technol. 2008, 19, 102–114. [Google Scholar] [CrossRef]
- Lugli, G.A.; Mangifesta, M.; Mancabelli, L.; Milani, C.; Turroni, F.; Viappiani, A.; van Sinderen, D.; Ventura, M. Compositional assessment of bacterial communities in probiotic supplements by means of metagenomic techniques. Int. J. Food Microbiol. 2019, 294, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Patro, J.N.; Ramachandran, P.; Barnaba, T.; Mammel, M.K.; Lewis, J.L.; Elkins, C.A. Culture-independent metagenomic surveillance of commercially available probiotics with high-throughput next-generation sequencing. MSphere 2016, 1, e00057-16. [Google Scholar] [CrossRef] [PubMed]
- Al-Otaibi, M. Evaluation of some probiotic fermented milk products from Al-Ahsa markets, Saudi Arabia. Am. J. Food Technol. 2009, 4, 1–8. [Google Scholar] [CrossRef]
- Morovic, W.; Hibberd, A.A.; Zabel, B.; Barrangou, R.; Stahl, B. Genotyping by PCR and High-Throughput Sequencing of Commercial Probiotic Products Reveals Composition Biases. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Patro, J.; Ramachandran, P.; Lewis, J.; Mammel, M.; Barnaba, T.; Pfeiler, E.; Elkins, C. Development and utility of the FDA ‘GutProbe’DNA microarray for identification, genotyping and metagenomic analysis of commercially available probiotics. J. Appl. Microbiol. 2015, 118, 1478–1488. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, e280. [Google Scholar] [CrossRef]
- Floch, M. Recommendations for probiotic use in humans—A 2014 update. Pharmaceuticals 2014, 7, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Rodighiero, V.; Celeste, T.; Rovetto, L.; De Vecchi, E. Microbiological evaluation of commercial probiotic products available in the USA in 2009. J. Chemother. 2010, 22, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Toscano, M.; Vecchi, E.D.; Rodighiero, V.; Drago, L. Microbiological and genetic identification of some probiotics proposed for medical use in 2011. J. Chemother. 2013, 25, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Dickson, E.M.; Riggio, M.P.; Macpherson, L. A novel species-specific PCR assay for identifying Lactobacillus fermentum. J. Med. Microbiol. 2005, 54, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; de’Angelis, G.L.; Shanahan, F. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Moura, P.; Simões, F.; Gírio, F.; Loureiro-Dias, M.C.; Esteves, M.P. PCR monitoring of Lactobacillus and Bifidobacterium dynamics in fermentations by piglet intestinal microbiota. J. Basic Microbiol. 2007, 47, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Usadel, B.; Lohse, M. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yuan, J.; Yiu, S.M.; Li, Z.; Xie, Y.; Chen, Y.; Shi, Y.; Zhang, H.; Li, Y.; Lam, T.W.; et al. COPE: An accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 2012, 28, 2870–2874. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Pevzner, P.A. Short read fragment assembly of bacterial genomes. Genome Res. 2008, 18, 324–330. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols; Springer: New York, NY, USA, 2000; pp. 365–386. [Google Scholar]
- Matsuki, T.; Watanabe, K.; Tanaka, R.; Fukuda, M.; Oyaizu, H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 1999, 65, 4506–4512. [Google Scholar] [PubMed]
- Torriani, S.; Zapparoli, G.; Dellaglio, F. Use of PCR-based methods for rapid differentiation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. Appl. Environ. Microbiol. 1999, 65, 4351–4356. [Google Scholar] [PubMed]
- Kok, R.G.; De Waal, A.; Schut, F.; Welling, G.W.; Weenk, G.; Hellingwerf, K.J. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 1996, 62, 3668–3672. [Google Scholar] [PubMed]
- Lick, S.; Keller, M.; Bockelmann, W.; Heller, J. Rapid identification of Streptococcus thermophilus by primer-specific PCR amplification based on its lacZ gene. Syst. Appl. Microbiol. 1996, 19, 74–77. [Google Scholar] [CrossRef]
- Walter, J.; Tannock, G.; Tilsala-Timisjarvi, A.; Rodtong, S.; Loach, D.; Munro, K.; Alatossava, T. Detection and identification of gastrointestinallactobacillus species by using denaturing gradient gel electrophoresis and species-specific pcr primers. Appl. Environ. Microbiol. 2000, 66, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Ahlroos, T.; Tynkkynen, S. Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J. Appl. Microbiol. 2009, 106, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Dave, R.; Shah, N. Evaluation of media for selective enumeration of Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactobacillus acidophilus, and bifidobacteria. J. Dairy Sci. 1996, 79, 1529–1536. [Google Scholar] [CrossRef]
- Tharmaraj, N.; Shah, N. Selective enumeration of Lactobacillus delbrueckii ssp. bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, bifidobacteria, Lactobacillus casei, Lactobacillus rhamnosus, and propionibacteria. J. Dairy Sci. 2003, 86, 2288–2296. [Google Scholar] [CrossRef]
| Bacteria | Medium | Growth Condition |
|---|---|---|
| Bifidobacterium spp. | MRS/ MRS-NPNL | anaerobic incubation at 37 °C for 72 h |
| Lactobacillus acidophilus | MRS-clindamycin | anaerobic incubation at 37 °C for 72 h |
| Lactobacillus casei | MRS-Bile | aerobic incubation at 37 °C for 48 h |
| Lactobacillus delbrueckii | MRS 5.2 | pH 5.2, anaerobic incubation at 45 °C for 72 h |
| Lactobacillus fermentum | MRS | anaerobic incubation at 37 °C for 48 h |
| Lactobacillus gasseri | MRS | anaerobic incubation at 37 °C for 48 h |
| Lactobacillus helveticus | MRS | aerobic incubation at 37 °C for 48 h |
| Lactobacillus paracasei | MRS-Bile | aerobic incubation at 37 °C for 48 h |
| Lactobacillus plantarum | MRS | anaerobic incubation at 37 °C for 48 h |
| Lactobacillus reuteri | MRS | aerobic incubation at 37 °C for 48 h |
| Lactobacillus rhamnosus | MRS Vancomycin | anaerobic incubation at 37 °C for 48 h |
| Lactobacillus salivarius | MRS | anaerobic incubation at 37 °C for 48 h |
| Streptococcus thermophilus | M17 Agar | aerobic incubation at 37 °C for 48 h |
| Lactococcus lactis | M17 Agar (0.5% glucose) | aerobic incubation at 30 °C for 48 h |
| Probiotic | Label Composition a | CFU Count ±SD b |
|---|---|---|
| PB2 | 8×109 CFU of L. acidophilus, B. animalis, B. longum, S. thermophilus | 8×109 ± 0.075 |
| PB4 | 9×109 CFU of L. rhamnosus, B. bifidum, B. animalis, L. acidophilus S. thermophilus, L. bulgaricus, B. longum | 7.5×109 ± 0.012 |
| PB5 | 2.0 × 109 CFU L. casei, L. plantarum | 2.0 × 109 ± 0.078 |
| PB6 | 30×109 B. animalis, 30×109 L. rhamnosus | 60 × 109 ± 0.080 |
| PB7 | L. acidophilus 5.3 × 109, B. longum 1 × 109 | 6.3 × 109 ± 0.0318 |
| PB8 | 6×109 CFU of L. acidophilus, L. fermentum, L. helveticus, L. paracasei, L. plantarum, L. rhamnosus | 5.0 × 109 ± 0.059 |
| PB9 | 17.5 × 109 CFU of B. longum, L. acidophilus, L. fermentum, L paracasei, L. plantarum, L. rhamnosus, S. thermophilis | 10.5 × 109 ± 0.013 |
| PB10 | 1×107 CFU of L. acidophilus, B. lactis, B. bifidum, L. planatarum | NG |
| PB11 | 1.425 × 109 CFU B. bifidum, 1.425 × 109 CFU B. longum (infantis), 9.6 × 109 CFU L. helviticus | 12.4 ×109 ± 0.005 |
| PB12 | 10 × 109 CFU of L. reuteri, L. salivarius, L. acidophilus, L. rhamnosus | 10 × 109 ± 0.033 |
| PB13 | 1x109 CFU of B. animalis | 1 × 109 ± 0.055 |
| PB14 | 6× 109 CFU of B. animalis, L. rhamnosus | 6× 109 ± 0.015 |
| PB15 | 5× 109 CFU of L. acidophilus, B. longum | 5× 109 ± 0.013 |
| PB16 | 4 × 109 CFU of L. acidophilus, B. longum, B. animalis, L. paracasei | 4 × 109 ± 0.071 |
| PB17 | 28 × 109 CFU of L. acidophilus, B. longum, B. bifidum, B. breve, B. lactis, L. bulgaricus, L. casei, L. helviticus, L. plantarum, L. reuteri, L. rhamnosus, L. salivarius, S. thermophilus, L. brevis, L. lactis | 16 × 109 ± 0.028 |
| PB18 | 4 × 109 CFU of L. gasseri, B. bifidum, B. longum | 4 × 109 ± 0.072 |
| PB19 | 3 × 109 of L. acidophilus, B. animalis, L. rhamnosus | 3 × 109 ± 0.063 |
| Sample | Raw Reads | Clean Reads | Raw Base(G) | Clean Base (G) | GC% |
|---|---|---|---|---|---|
| PB2 | 19,956,035 | 5,410,890 | 5.99 | 1.62 | 44.66 |
| PB4 | 8,399,739 | 6,419,867 | 2.52 | 1.93 | 46.55 |
| PB5 | 9,661,983 | 7,548,271 | 2.9 | 2.26 | 46.39 |
| PB6 | 9,194,286 | 7,793,312 | 2.76 | 2.34 | 48.39 |
| PB7 | 8,835,931 | 6,599,652 | 2.65 | 1.98 | 43.4 |
| PB8 | 8,193,811 | 5,031,234 | 2.46 | 1.51 | 46.44 |
| PB9 | 7,913,048 | 6,447,026 | 2.37 | 1.93 | 50 |
| PB10 | 9,669,794 | 6,868,151 | 2.9 | 2.06 | 45.11 |
| PB11 | 9,486,662 | 7,029,632 | 2.85 | 2.11 | 42.34 |
| PB12 | 11,201,502 | 8,544,999 | 3.36 | 2.56 | 39.22 |
| PB13 | 13,010,708 | 11,301,642 | 3.9 | 3.39 | 57.88 |
| PB14 | 13,461,984 | 9,517,061 | 4.04 | 2.85 | 43.04 |
| PB15 | 9,865,374 | 7,615,037 | 2.96 | 2.29 | 46.2 |
| PB16 | 8,262,088 | 2,411,023 | 2.48 | 0.72 | 52.7 |
| PB17 | 10,252,995 | 8,199,392 | 3.08 | 2.46 | 46.2 |
| PB18 | 7,823,958 | 5,023,448 | 2.35 | 1.51 | 42.55 |
| PB19 | 8,257,723 | 6,415,206 | 2.48 | 1.93 | 37.89 |
| Bacterial Contents Mentioned on the Product Label | Bacterial Contents Identified during Sequence Analysis | |
|---|---|---|
| PB2 | Not Mentioned | Lactobacillus rhamnosus LOCK908 |
| Not Mentioned | Lactobacillus casei W56 | |
| Not Mentioned | Lactobacillus plantarum strain B21 | |
| Lactobacillus acidophilus strain FS14 | Lactobacillus acidophilus strain FSI4 | |
| PB4 | Lactobacillus rhamnosus ATCC 53103 | Lactobacillus rhamnosus ATCC 53103 |
| Bifidobacterium bifidum ATCC 29521 | Bifidobacterium bifidum ATCC 29521 | |
| Lactobacillus acidophilus strain FSI4 | Lactobacillus acidophilus strain FSI4 | |
| Bifidobacterium animalis | Bifidobacterium animalis strain A6 | |
| Bifidobacterium longum | Bifidobacterium longum subsp. infantis ATCC 15697 | |
| PB5 | Lactobacillus casei | Lactobacillus casei W56 |
| Lactobacillus plantarum ZS2058 | Lactobacillus plantarum ZS2058 | |
| PB6 | Lactobacillus rhamnosus LOCK908 | Lactobacillus rhamnosus LOCK908 |
| Bifidobacterium animalis sbsp. lactis | Bifidobacterium animalis sbsp. lactis strain A6 | |
| PB7 | Not Mentioned | Lactobacillus casei BL23 |
| Lactobacillus acidophilus La-14 | Lactobacillus acidophilus La-14 | |
| PB8 | Lactobacillus paracasei | Lactobacillus paracasei strain L9 |
| Lactobacillus rhamnosus LOCK908 | Lactobacillus rhamnosus LOCK908 | |
| Lactobacillus plantarum subsp. plantarum ST-III | Lactobacillus plantarum subsp. plantarum ST-III | |
| Lactobacillus helveticus R0052 | Lactobacillus helveticus R0052 | |
| Lactobacillus fermentum 3872 | Lactobacillus fermentum 3872 | |
| PB9 | Lactobacillus rhamnosus ATCC 53103 | Lactobacillus rhamnosus ATCC 53103 |
| Lactobacillus plantarum LZ95 | Lactobacillus plantarum strain LZ95 | |
| Lactobacillus paracasei L9 | Lactobacillus paracasei strain L9 | |
| Lactobacillus fermentum 3872 | Lactobacillus fermentum 3872 | |
| PB10 | Lactobacillus plantarum B21 | Lactobacillus plantarum strain B21 |
| Bifidobacterium bifidum | Bifidobacterium bifidum ATCC 29521 | |
| Lactobacillus acidophilus La-14 | Lactobacillus acidophilus La-14 | |
| Bifidobacterium animalis subsp lactis | Bifidobacterium animalis strain A6 | |
| PB11 | Lactobacillus helveticus R0052 | Lactobacillus helveticus R0052 |
| Bifidobacterium bifidum BGN4 | Bifidobacterium bifidum BGN4 | |
| Bifidobacterium infantis subsp. infantis | Bifidobacterium longum subsp. infantis ATCC 15697 | |
| PB12 | Lactobacillus rhamnosus 53103 | Lactobacillus rhamnosus ATCC 53103 |
| Lactobacillus reuteri | Lactobacillus reuteri SD2112 | |
| Lactobacillus acidophilus | Lactobacillus acidophilus strain FSI4 | |
| Lactobacillus salivarius | Lactobacillus salivarius strain Ren | |
| PB13 | Bifidobacterium animalis | Bifidobacterium animalis strain A6 |
| PB14 | Not Mentioned | Lactobacillus helveticus R0052 |
| Not Mentioned | Bifidobacterium longum subsp. infantis 157F | |
| PB15 | Not Mentioned | Lactobacillus rhamnosus ATCC 53103 |
| Not Mentioned | Lactobacillus plantarum strain B21 | |
| PB16 | Lactobacillus paracasei L9 | Lactobacillus paracasei strain L9 |
| Bifidobacterium longum subsp. longum | Bifidobacterium longum subsp. longum JCM 1217 | |
| Lactobacillus acidophilus strain FS14 | Lactobacillus acidophilus strain FSI4 | |
| Bifidobacterium animalis subsp. lactis | Bifidobacterium animalis subsp. lactis strain A6 | |
| PB17 | Lactobacillus rhamnosus LOCK908 | Lactobacillus rhamnosus LOCK908 |
| Bifidobacterium longum subsp. longum | Bifidobacterium longum subsp. longum JCM 1217 | |
| Lactobacillus plantarum subsp. plantarum | Lactobacillus plantarum subsp. plantarum ST-III | |
| Lactobacillus helveticus | Lactobacillus helveticus DPC 4571 | |
| Lactococcus lactis subsp. lactis | Lactococcus lactis subsp. lactis CV56 | |
| Lactobacillus acidophilus FSI4 | Lactobacillus acidophilus strain FSI4 | |
| Lactobacillus casei subsp. casei | Lactobacillus casei subsp. casei ATCC 393 | |
| Lactobacillus delbrueckii subsp. bulgaricus | Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 | |
| Bifidobacterium breve | Bifidobacterium breve JCM 7017 | |
| Lactobacillus reuteri | Lactobacillus reuteri JCM 1112 | |
| Streptococcus thermophilus | Streptococcus thermophilus LMD-9 | |
| Bifidobacterium bifidum BGN4 | Bifidobacterium bifidum BGN4 | |
| Bifidobacterium animalis subsp. lactis | Bifidobacterium animalis strain A6 | |
| Lactobacillus brevis | Lactobacillus brevis KB290 | |
| Lactobacillus salivarius | Lactobacillus salivarius strain JCM1046 | |
| PB18 | Bifidobacterium bifidum | Bifidobacterium bifidum S17 |
| Bifidobacterium longum | Bifidobacterium longum strain: 105-A | |
| Lactobacillus gasseri | Lactobacillus gasseri ATCC 33323 | |
| PB19 | Lactobacillus rhamnosus LOCK908 | Lactobacillus rhamnosus LOCK908 |
| Lactobacillus acidophilus strain FSI4 | Lactobacillus acidophilus strain FSI4 | |
| Bifidobacterium animalis subsp. lactis | Bifidobacterium animalis strain A6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, M.; Raza, A.; Ye, L.; Yu, Z. Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing. Microorganisms 2019, 7, 188. https://doi.org/10.3390/microorganisms7070188
Ullah M, Raza A, Ye L, Yu Z. Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing. Microorganisms. 2019; 7(7):188. https://doi.org/10.3390/microorganisms7070188
Chicago/Turabian StyleUllah, Mati, Ali Raza, Li Ye, and Zhu Yu. 2019. "Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing" Microorganisms 7, no. 7: 188. https://doi.org/10.3390/microorganisms7070188
APA StyleUllah, M., Raza, A., Ye, L., & Yu, Z. (2019). Viability and Composition Validation of Commercial Probiotic Products by Selective Culturing Combined with Next-Generation Sequencing. Microorganisms, 7(7), 188. https://doi.org/10.3390/microorganisms7070188






