Microbial and Functional Profile of the Ceca from Laying Hens Affected by Feeding Prebiotics, Probiotics, and Synbiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals and Experimental Diets
2.3. Cecal Sample Collection and DNA Extraction
2.4. 16S rRNA Gene Amplification, Sequencing, and Analysis
2.5. Metagenome Sequencing and Analysis
3. Results
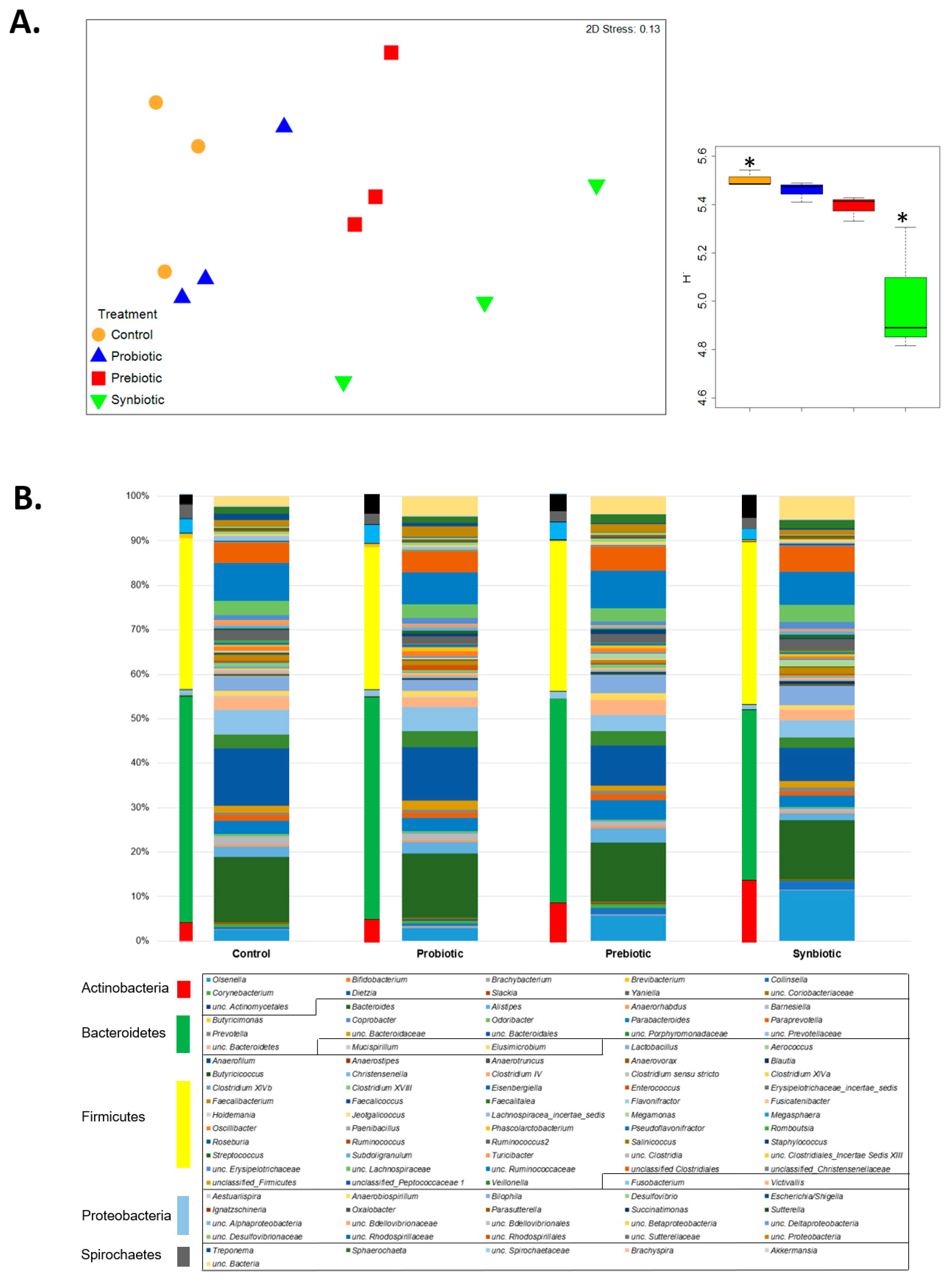
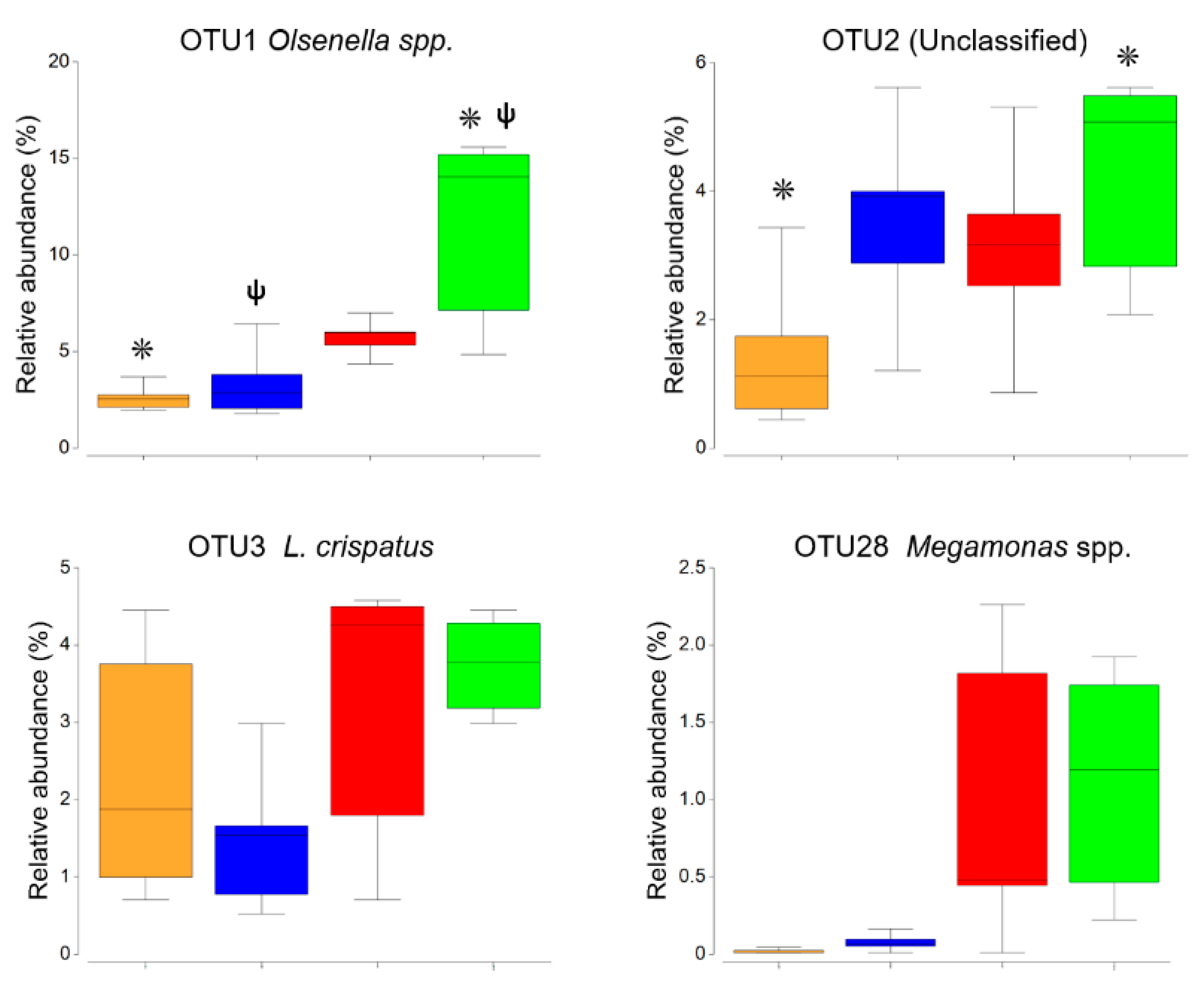
3.1. Microbial Community Analysis Based on 16S rRNA Gene Amplicon Sequencing
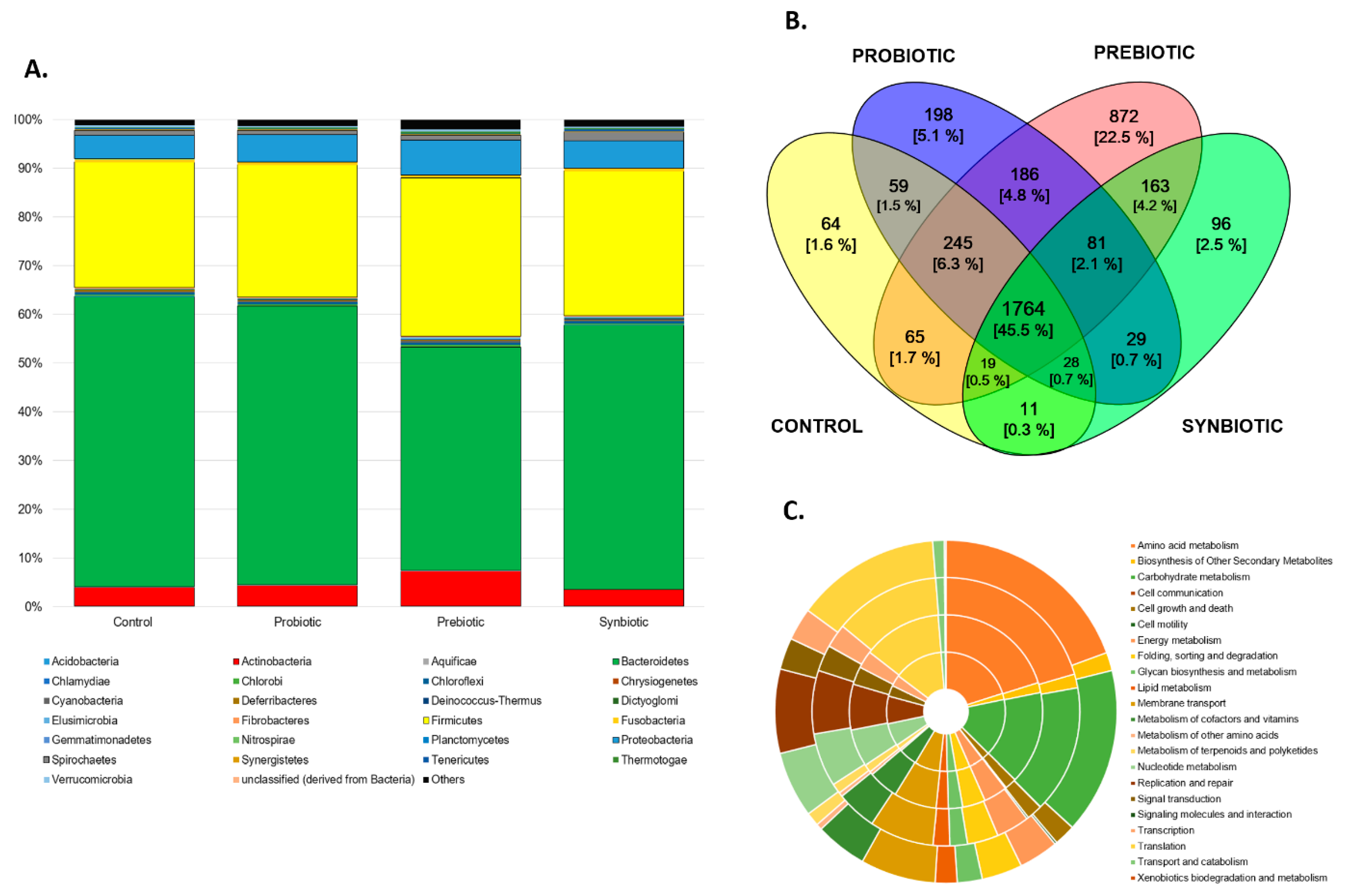
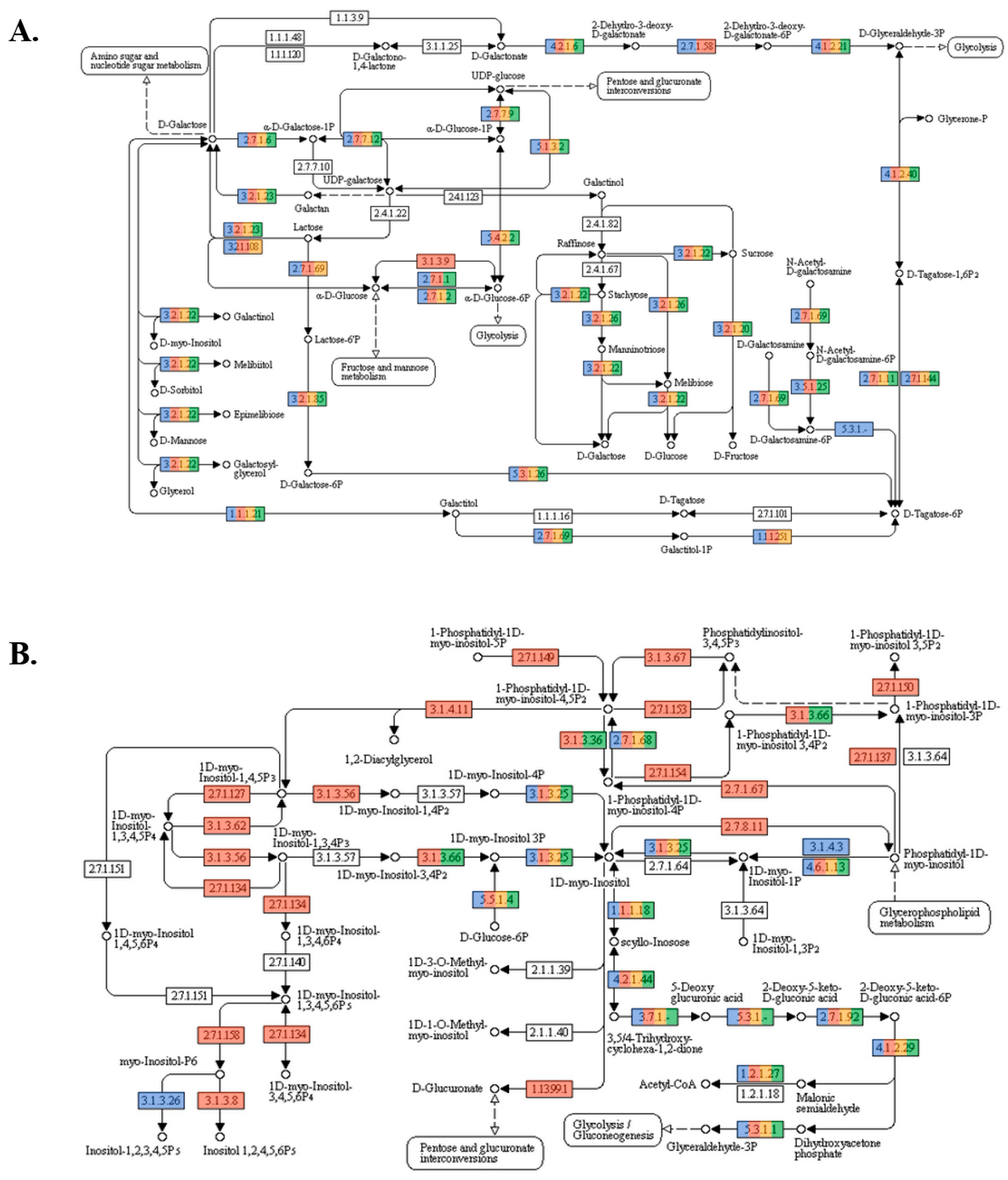
3.2. Metagenomic Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef]
- Borda-Molina, D.; Zuber, T.; Siegert, W.; Camarinha-Silva, A.; Feuerstein, D.; Rodehutscord, M. Effects of protease and phytase supplements on small intestinal microbiota and amino acid digestibility in broiler chickens. Poult. Sci. 2019. [Google Scholar] [CrossRef]
- Pineda-Quiroga, C.; Camarinha-Silva, A.; Borda-Molina, D.; Atxaerandio, R.; Ruiz, R.; García-Rodríguez, A. Feeding broilers with dry whey powder and whey protein concentrate affected productive performance, ileal digestibility of nutrients and cecal microbiota community. Animal 2018, 12, 692–700. [Google Scholar] [CrossRef]
- Angelakis, E. Weight gain by gut microbiota manipulation in productive animals. Microb. Pathog. 2017, 106, 162–170. [Google Scholar] [CrossRef]
- Pourabedin, M.; Zhao, X. Prebiotics and gut microbiota in chickens. FEMS Microbiol. Lett. 2015, 362, fnv122. [Google Scholar] [CrossRef]
- Scott, K.P.; Antoine, J.-M.; Midtvedt, T.; van Hemert, S. Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 2015, 26, 25877. [Google Scholar] [CrossRef]
- Awad, W.A.; Ghareeb, K.; Abdel-Raheem, S.; Böhm, J. Effects of dietary inclusion of probiotic and synbiotic on growth performance, organ weights, and intestinal histomorphology of broiler chickens. Poult. Sci. 2009, 88, 49–56. [Google Scholar] [CrossRef]
- De Maesschalck, C.; Eeckhaut, V.; Maertens, L.; de Lange, L.; Marchal, L.; Nezer, C.; de Baere, S.; Croubels, S.; Daube, G.; Dewulf, J.; et al. Effects of Xylo-Oligosaccharides on Broiler Chicken Performance and Microbiota. Appl. Environ. Microbiol. 2015, 81, 5880–5888. [Google Scholar] [CrossRef]
- Ocejo, M.; Oporto, B.; Hurtado, A. 16S rRNA amplicon sequencing characterization of cecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci. Rep. 2019, 9, 2506. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef]
- Jozefiak, D.; Rutkowski, A.; Kaczmarek, S.; Jensen, B.B.; Engberg, R.M.; Højberg, O. Effect of β-glucanase and xylanase supplementation of barley- and rye-based diets on cecal microbiota of broiler chickens. Br. Poult. Sci. 2010, 51, 546–557. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Qu, A.; Brulc, J.M.; Wilson, M.K.; Law, B.F.; Theoret, J.R.; Joens, L.A.; Konkel, M.E.; Angly, F.; Dinsdale, E.A.; Edwards, R.A.; et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 2008, 3, e2945. [Google Scholar] [CrossRef]
- Clavel, T.; Lagkouvardos, I.; Hiergeist, A. Microbiome sequencing: Challenges and opportunities for molecular medicine. Expert Rev. Mol. Diagn. 2016, 16, 795–805. [Google Scholar] [CrossRef]
- Kohl, K.D. Diversity and function of the avian gut microbiota. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2012, 182, 591–602. [Google Scholar] [CrossRef]
- Dinsdale, E.A.; Edwards, R.A.; Hall, D.; Angly, F.; Breitbart, M.; Brulc, J.M.; Furlan, M.; Desnues, C.; Haynes, M.; Li, L.; et al. Functional metagenomic profiling of nine biomes. Nature 2008, 452, 629–632. [Google Scholar] [CrossRef]
- Yoon, S.S.; Kim, E.-K.; Lee, W.-J. Functional genomic and metagenomic approaches to understanding gut microbiota-animal mutualism. Curr. Opin. Microbiol. 2015, 24, 38–46. [Google Scholar] [CrossRef]
- Pineda-Quiroga, C.; Atxaerandio, R.; Zubiria, I.; Gonzalez-Pozuelo, I.; Hurtado, A.; Ruiz, R.; Garcia-Rodriguez, A. Productive performance and cecal microbial counts of floor housed laying hens supplemented with dry whey powder alone or combined with Pediococcus acidilactici in the late phase of production. Livestock Sci. 2017, 195, 9–12. [Google Scholar] [CrossRef]
- Kaewtapee, C.; Burbach, K.; Tomforde, G.; Hartinger, T.; Camarinha-Silva, A.; Heinritz, S.; Seifert, J.; Wiltafsky, M.; Mosenthin, R.; Rosenfelder-Kuon, P. Effect of Bacillus subtilis and Bacillus licheniformis supplementation in diets with low- and high-protein content on ileal crude protein and amino acid digestibility and intestinal microbiota composition of growing pigs. J. Anim. Sci. Biotechnol. 2017, 8, 37. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities. An Approach to Statistical Analysis and Interpretation; Plymouth Marine Laboratory: Plymouth, UK, 1997; ISBN 9781855311404. [Google Scholar]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Gao, P.; Ma, C.; Sun, Z.; Wang, L.; Huang, S.; Su, X.; Xu, J.; Zhang, H. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 2017, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Chalvatzi, S.; Kalamaki, M.S.; Arsenos, G.; Fortomaris, P. Dietary supplementation with the clay mineral palygorskite affects performance and beneficially modulates cecal microbiota in laying pullets. J. Appl. Microbiol. 2016, 120, 1033–1040. [Google Scholar] [CrossRef]
- Rezvani, M.; Mendoza, M.; Koci, M.D.; Daron, C.; Levy, J.; Hassan, H.M. Draft Genome Sequence of Lactobacillus crispatus C25 Isolated from Chicken Cecum. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Neal-McKinney, J.M.; Lu, X.; Duong, T.; Larson, C.L.; Call, D.R.; Shah, D.H.; Konkel, M.E. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS ONE 2012, 7, e43928. [Google Scholar] [CrossRef]
- Ludwig, W.; Schleifer, K.-H.; Whitman, W.B. Revised road map to the phylum Firmicutes. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Garrity, G.M., Ed.; Springer: New York, NY, USA, 2001; pp. 1–13. ISBN 978-0-387-95041-9. [Google Scholar]
- Bennett, D.C.; Tun, H.M.; Kim, J.E.; Leung, F.C.; Cheng, K.M. Characterization of cecal microbiota of the emu (Dromaius novaehollandiae). Vet. Microbiol. 2013, 166, 304–310. [Google Scholar] [CrossRef]
- Segerman, B. The genetic integrity of bacterial species: The core genome and the accessory genome, two different stories. Front. Cell. Infect. Microbiol. 2012, 2. [Google Scholar] [CrossRef]
- SVIHUS, B.; Choct, M.; Classen, H.L. Function and nutritional roles of the avian ceca: A review. Worlds Poult. Sci. J. 2013, 69, 249–264. [Google Scholar] [CrossRef]
- Annison, E.F.; Hill, K.J.; Kenworthy, R. Volatile fatty acids in the digestive tract of the fowl. Br. J. Nutr. 1968, 22, 207–216. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef]
- Marounek, M.; Skřivan, M.; Rosero, O.; Rop, O. Intestinal and total tract phytate digestibility and phytase activity in the digestive tract of hens fed a wheat-maize-soyabean diet. J. Anim. Feed Sci. 2010, 19, 430–439. [Google Scholar] [CrossRef]
- Rodehutscord, M.; Rosenfelder, P. Update on phytate degradation pattern in the gastrointestinal tract of pigs and broiler chickens. Chapter 1. In Phytate Destruction—Consequences for Precision in Animal Nutrition; Wageningen Academic Publishers: Wageningen, The Netherlands, 2016. [Google Scholar]
- Waldroup, P.W.; Kersey, J.H.; Saleh, E.A.; Fritts, C.A.; Yan, F.; Stilborn, H.L.; Crum, R.C.; Raboy, V. Nonphytate phosphorus requirement and phosphorus excretion of broiler chicks fed diets composed of normal or high available phosphate corn with and without microbial phytase. Poult. Sci. 2000, 79, 1451–1459. [Google Scholar] [CrossRef]
- Ahmadi, H.; Rodehutscord, M. A meta-analysis of responses to dietary nonphytate phosphorus and phytase in laying hens. Poult. Sci. 2012, 91, 2072–2078. [Google Scholar] [CrossRef]
- Brown, K.; Uwiera, R.R.E.; Kalmokoff, M.L.; Brooks, S.P.J.; Inglis, G.D. Antimicrobial growth promoter use in livestock: A requirement to understand their modes of action to develop effective alternatives. Int. J. Antimicrob. Agents 2017, 49, 12–24. [Google Scholar] [CrossRef]
- White, D.G.; Ayers, S.; Maurer, J.J.; Thayer, S.G.; Hofacre, C. Antimicrobial Susceptibilities of Staphylococcus aureus Isolated from Commercial Broilers in Northeastern Georgia. Avian Dis. 2003, 47, 203–210. [Google Scholar] [CrossRef]
| Statistical Differences between Diets Based on the PERMANOVA Results | |||
|---|---|---|---|
| Diets 1 | Groups | t-Value | p (perm) |
| Control | Probiotic | 12.865 | 0.106 |
| Prebiotic | 1.657 | 0.001 | |
| Synbiotic | 16.105 | 0.001 | |
| Probiotic | Prebiotic | 16.325 | 0.001 |
| Synbiotic | 14.991 | 0.001 | |
| Prebiotic | Synbiotic | 1.225 | 0.190 |
| Function | Control vs. Probiotic | Control vs. Prebiotic | Control vs. Synbiotic | Prebiotic vs. Probiotic | Prebiotic vs. Synbiotic | Probiotic vs. Synbiotic | |
|---|---|---|---|---|---|---|---|
| Starch and sucrose metabolism | Beta-phosphoglucomutase [K01838] | 1.64 | 1.72 | 2.33 | |||
| Sucrose phosphorylase [K00690] | −1.82 | ||||||
| Maltose phosphorylase [K00691] | −1.11 | ||||||
| Maltose-6’-phosphate glucosidase [K01232] | −1.03 | ||||||
| Beta-phosphoglucomutase [K01838] | −2.34 | ||||||
| Cyclomaltodextrinase [K01208] | −1.23 | −2.32 | |||||
| Glucan endo-1,3-beta-d-glucosidase [K01199] | −1.69 | −3.05 | −2.11 | ||||
| Cellulose synthase (UDP-forming) [K00694] | −2.37 | ||||||
| Glycolisis/gluconeogenesis | Pyruvate decarboxylase [K01568] | −1.1 | −2.12 | ||||
| Acetyl-CoA synthetase (ADP-forming) [K01905] | −2.1 | −1.47 | −3.17 | ||||
| Piruvate metabolism | D-lactate dehydrogenase (cytochrome) [K00102] | 3.48 | 4.13 | 3.69 | |||
| Citrate cycle (TCA cycle) | 2-methylisocitrate dehydratase [K01682] | 1.4 | 2.05 | 1.84 | |||
| Glycerophospholipid metabolism | Glycerol-3-phosphate dehydrogenase subunit B [K00112] | 1.88 | 1.12 | 2.69 | |||
| Glycine, serine and threonine metabolism | Glycine amidinotransferase [K00613] | 1.38 | 1.88 | 2.56 | |||
| Cystathionine gamma-lyase [K01758] | 1.63 | 1.63 | 2.56 | ||||
| Glycine N-methyltransferase [K00552] | Not in Ctrl | −1 | Not in Syn | Not in Prob | Not in Syn | ||
| Creatinase [K08688] | −1 | −1 | Not in Syn | ||||
| Diaminobutyric acid acetyltransferase[K06718] | −1 | −1 | Not in Syn | ||||
| Cetaine-homocysteine S-methyltransferase [K00544] | Not in Prob | −1 | Not in Syn | ||||
| Choline dehydrogenase [K00108] | −2.11 | 1.23 | 3.81 | ||||
| Arginine and proline metabolism | Creatinine amidohydrolase [K01470] | 1.2 | 1.44 | 1.52 | |||
| Ornithine decarboxilase [K01581] | 1.8 | 1.22 | 1.89 | ||||
| Cysteine metabolism | Aromatic-amino-acid transaminase [K00832] | 1.21 | 1.37 | 2.18 | |||
| Cysteine and methionine metabolism | Homocysteine S-methyltransferase [K00547] | −1.76 | |||||
| Aromatic-amino-acid transaminase [K00832] | −2.18 | ||||||
| (R)-2-hydroxyacid dehydrogenase [K05884] | −1.69 | −1.05 | |||||
| Tyrosine metabolism | 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase [K05921] | 1.99 | 1.22 | 2.1 | |||
| Pantothenate and CoA biosynthesis | Type I pantothenate kinase [K00867] | 1.02 | 1.2 | 1.68 | |||
| Phosphopantothenoylcysteine decarboxylase [K01598] | 1.79 | 1.8 | 1.1 | ||||
| Folate biosynthesis | Para-aminobenzoate synthetase [K01247] | 3.57 | 3.72 | 2.1 | |||
| Riboflavin metabolism | Low molecular weight phosphotyrosine protein Phosphatase [K14394] | 1.31 | 1.22 | 3.1 | |||
| Lysine degradation | 5-aminovalerate aminotransferase [K14268] | Not in Prob | Not in Preb | Not in Syn | |||
| Glutarate semialdehyde dehydrogenase [K14269] | Not in Prob | Not in Preb | Not in Syn | ||||
| β-lactam resistance | Methicillin resistance protein [K02547] | −2.82 | −2.17 | Not in Syn | |||
| Butanoate metabolism | Glutaconate CoA-transferase, subunit A [K01039] | −1.4 | Not in Prob | 2.62 | |||
| 4-hydroxybutyrate dehydrogenase [K00043] | / | 1.09 | 2.44 | ||||
| 3-hydroxybutyrate dehydrogenase [K00019] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| Butanediol dehydrogenase/diacetyl reductase [K00004] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| Propanoate metabolism | 1-aminocyclopropane-1-carboxylate deaminase [K01505] | −1.22 | / | 1.8 | |||
| 2-methylcitrate dehydratase [K05608] | −1.57 | 1.61 | Not in Syn | ||||
| 1-aminocyclopropane-1-carboxylate deaminase [K00923] | −1.22 | / | 1.7 | ||||
| 2-methylcitrate synthase [K01659] | / | / | 1.73 | ||||
| Methylisocitrate lyase [K03417] | / | / | 2.44 | ||||
| Fructose and mannose metabolism | mannitol 2-dehydrogenase [K00045] | / | / | 1.19 | |||
| PFK; 6-phosphofructo-2-kinase [K00900] | Not in Ctrl | 1.01 | 1.7 | ||||
| Fructan beta-fructosidase [K03332] | −1.63 | 1.32 | Not in Syn | ||||
| Inositol phosphate metabolism | 5-dehydro-2-deoxygluconokinase [K03338] | −1.06 | 1.2 | Not in Syn | |||
| iolB; 5-deoxy-glucuronate isomerase [K03337] | Not in Ctrl | 1.76 | 1.7 | ||||
| Galactose metabolism | Lactase-phlorizin hydrolase [K01229] | −3.22 | 2.23 | Not in Syn | |||
| Galactitol-1-phosphate 5-dehydrogenase [K00094] | −1.37 | 2.38 | Not in Syn | ||||
| Tagatose 6-phosphate kinase [K00917] | / | / | 1.17 | ||||
| 2-dehydro-3-deoxyphosphogalactonate aldolase [K01631] | / | 2.97 | 1.12 | ||||
| Galactonate dehydratase [K01684] | / | / | 1.35 | ||||
| Maltase-glucoamylase [K12047] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| N-acetylgalactosamine-6-phosphate deacetylase [K02079] | Not in Ctrl | Not in Preb | Not in Prob | ||||
| Phenylalanine, tyrosine and tryptophan biosynthesis | Anthranilate synthase/phosphoribosyltransferase [K13497] | −4.96 | 1.8 | / | |||
| Indole-3-glycerol phosphate synthase/Phosphoribosylanthranilate isomerase [K13498] | −1.42 | 1.26 | 1.26 | ||||
| Shikimate kinase/3-dehydroquinate synthase [K13829] | −1.09 | 1.78 | 2.99 | ||||
| Chorismate mutase/prephenate dehydratase [K14170] | / | / | 1.27 | ||||
| Cyclohexadienyl dehydratase [K01713] | Not in Ctrl | Not in Preb | Not in Prob | ||||
| Fatty acid metabolism | Long-chain-fatty-acid-[acyl-carrier-protein] Ligase [K05939] | −2.17 | 1.55 | / | |||
| Cytochrome P450, family 4, subfamily A [K07425] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| Carnitine O-palmitoyltransferase 2 [K08766] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| Fatty acid biosynthesis | Fatty acid synthase, bacteria type [K11533] | / | 1.22 | 4.56 | |||
| Oleoyl-[acyl-carrier-protein] hydrolase [K01071] | −1.7 | / | −1.24 | ||||
| Thiamine metabolism | Hydroxyethylthiazole kinase [K14154] | / | 2.23 | 1.11 | |||
| Thiamine-phosphate diphosphorylase [K14153] | −1.22 | / | Not in Syn | ||||
| Retinol metabolism | Retinol dehydrogenase 16 [K11154] | Not in Ctrl | Not in Prob | Not in Syn | |||
| Diacylglycerol O-acyltransferase 2-like protein 4 [K11156] | Not in Ctrl | Not in Prob | Not in Syn | ||||
| All-trans-retinol 13,14-reductase [K09516] | −1.1 | −1.76 | −1.83 | ||||
| Ascorbate and aldarate metabolism | D-threo-aldose 1-dehydrogenase [K00064] | −1.23 | / | 2.13 | |||
| l-ribulose-5-phosphate 3-epimerase [K03079] | −1.1 | −3.05 | 1.58 | ||||
| Glyoxylate and dicarboxylate metabolism | Formate dehydrogenase [K00122] | Not in Ctrl | −2.94 | / | |||
| Oxalyl-CoA decarboxylase [K01577] | / | −2.4 | 1.07 | ||||
| N,N-dimethylformamidase [K03418] | Not in Ctrl | Not in Preb | Not in Syn | ||||
| Formate dehydrogenase-N, gamma subunit [K04509] | Not in Ctrl | Not in Preb | Not in Syn | ||||
| Glycolate oxidase FAD binding subunit | Not in Ctrl | Not in Preb | Not in Syn | ||||
| Oxalate decarboxylase [K01569] | Not in Ctrl | Not in Preb | Not in Syn | ||||
| Malate dehydrogenase [K00025] | Not in Ctrl | −0.15 | Not in Syn | ||||
| Alanine, aspartate and glutamate metabolism | Aspartate 4-decarboxylase [K09758] | / | −1.17 | 2.61 | |||
| 1-pyrroline-5-carboxylate dehydrogenase [K00294] | / | / | 1.01 | ||||
| Carbamoyl-phosphate synthase [K01954] | / | −1.27 | 1.97 | ||||
| Delta 1-pyrroline-5-carboxylate dehydrogenase [K13821] | −1.59 | / | 2.3 | ||||
| Phenylalanine metabolism | Phenylacetaldehyde dehydrogenase [K00146] | −2.47 | −2.94 | / | |||
| 2-keto-4-pentenoate hydratase [K02554] | −1.57 | −1.94 | / | ||||
| Cinnamic acid dioxygenase subunit alpha [K05708] | −1.31 | −2.67 | / | ||||
| Nicotinate and nicotinamide metabolism | UDP-sugar diphosphatase [K11751] | −1.49 | / | / | |||
| 5′-nucleotidase [K01081] | / | −1.05 | / | ||||
| NAD(P) transhydrogenase [K00322] | / | −1.81 | / | ||||
| Nicotinamide-nucleotide adenylyltransferase [K00952] | / | −3.16 | / | ||||
| Purine nucleosidase [K01239] | −1.08 | / | / | ||||
| NAD(P) transhydrogenase subunit alpha [K00324] | −1.1 | / | / | ||||
| Amino sugar and nucleotide sugar metabolism | Chitin deacetylase [K01452] | / | −2.47 | −2.12 | |||
| Valine, leucine and isoleucine biosynthesis | Valine—pyruvate aminotransferase [K00835] | −3.69 | −4.05 | / | |||
| Steroid hormone biosynthesis | Steroid delta-isomerase [K01822] | / | −1.68 | −1.17 | |||
| Steryl-sulfatase [K01131] | −1.1 | −2.45 | −2.12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pineda-Quiroga, C.; Borda-Molina, D.; Chaves-Moreno, D.; Ruiz, R.; Atxaerandio, R.; Camarinha-Silva, A.; García-Rodríguez, A. Microbial and Functional Profile of the Ceca from Laying Hens Affected by Feeding Prebiotics, Probiotics, and Synbiotics. Microorganisms 2019, 7, 123. https://doi.org/10.3390/microorganisms7050123
Pineda-Quiroga C, Borda-Molina D, Chaves-Moreno D, Ruiz R, Atxaerandio R, Camarinha-Silva A, García-Rodríguez A. Microbial and Functional Profile of the Ceca from Laying Hens Affected by Feeding Prebiotics, Probiotics, and Synbiotics. Microorganisms. 2019; 7(5):123. https://doi.org/10.3390/microorganisms7050123
Chicago/Turabian StylePineda-Quiroga, Carolina, Daniel Borda-Molina, Diego Chaves-Moreno, Roberto Ruiz, Raquel Atxaerandio, Amélia Camarinha-Silva, and Aser García-Rodríguez. 2019. "Microbial and Functional Profile of the Ceca from Laying Hens Affected by Feeding Prebiotics, Probiotics, and Synbiotics" Microorganisms 7, no. 5: 123. https://doi.org/10.3390/microorganisms7050123
APA StylePineda-Quiroga, C., Borda-Molina, D., Chaves-Moreno, D., Ruiz, R., Atxaerandio, R., Camarinha-Silva, A., & García-Rodríguez, A. (2019). Microbial and Functional Profile of the Ceca from Laying Hens Affected by Feeding Prebiotics, Probiotics, and Synbiotics. Microorganisms, 7(5), 123. https://doi.org/10.3390/microorganisms7050123





