Mycobacterium smegmatis But Not Mycobacterium avium subsp. hominissuis Causes Increased Expression of the Long Non-Coding RNA MEG3 in THP-1-Derived Human Macrophages and Associated Decrease of TGF-β
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Cell Culture
2.3. Infection Experiments
2.4. Expression Analysis Using RT-qPCR
2.5. Statistical Analysis
3. Results
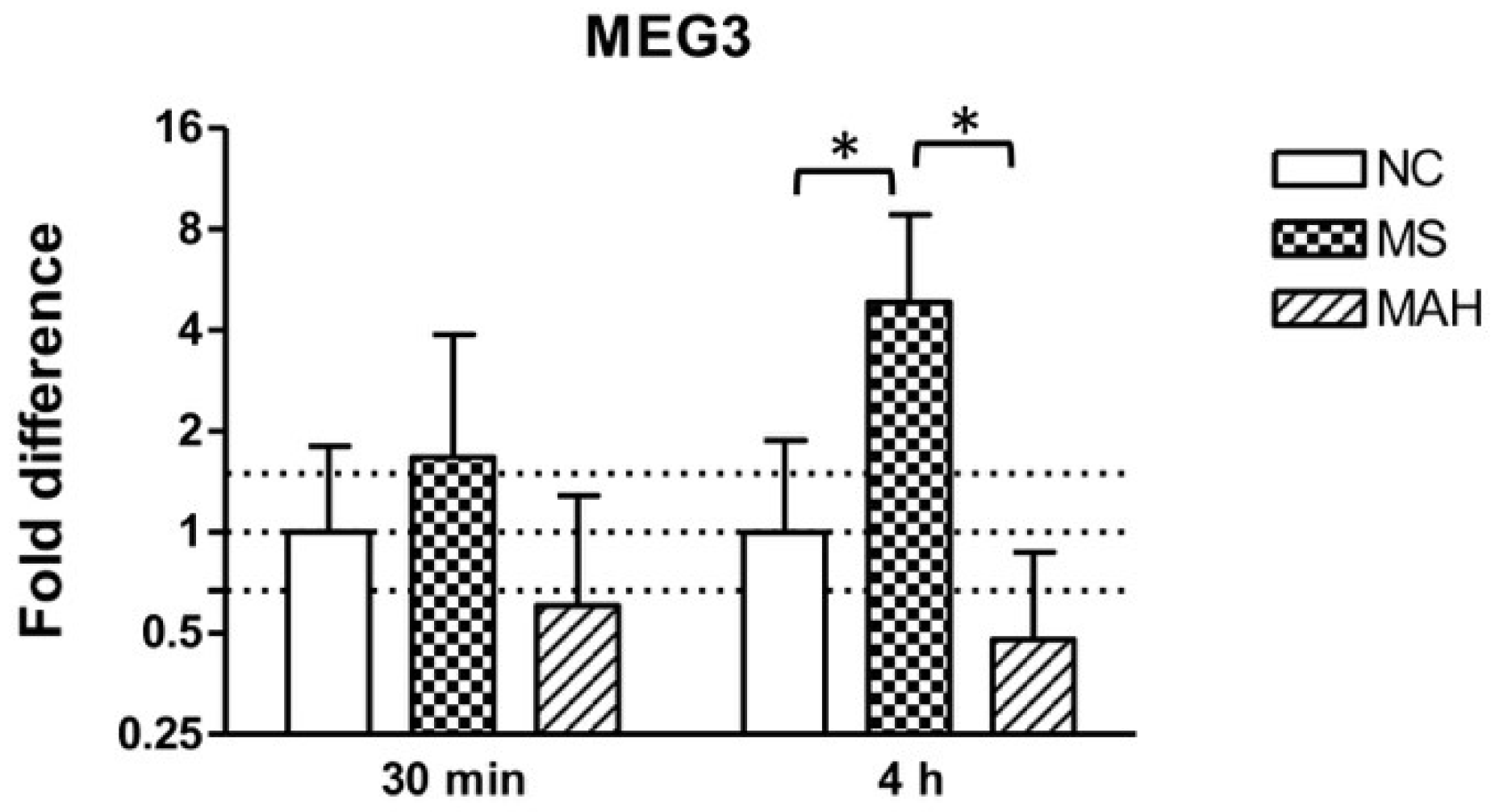
3.1. Infection with M. smegmatis But Not M. avium subsp. hominissuis Leads to Upregulation of the lncRNA MEG3
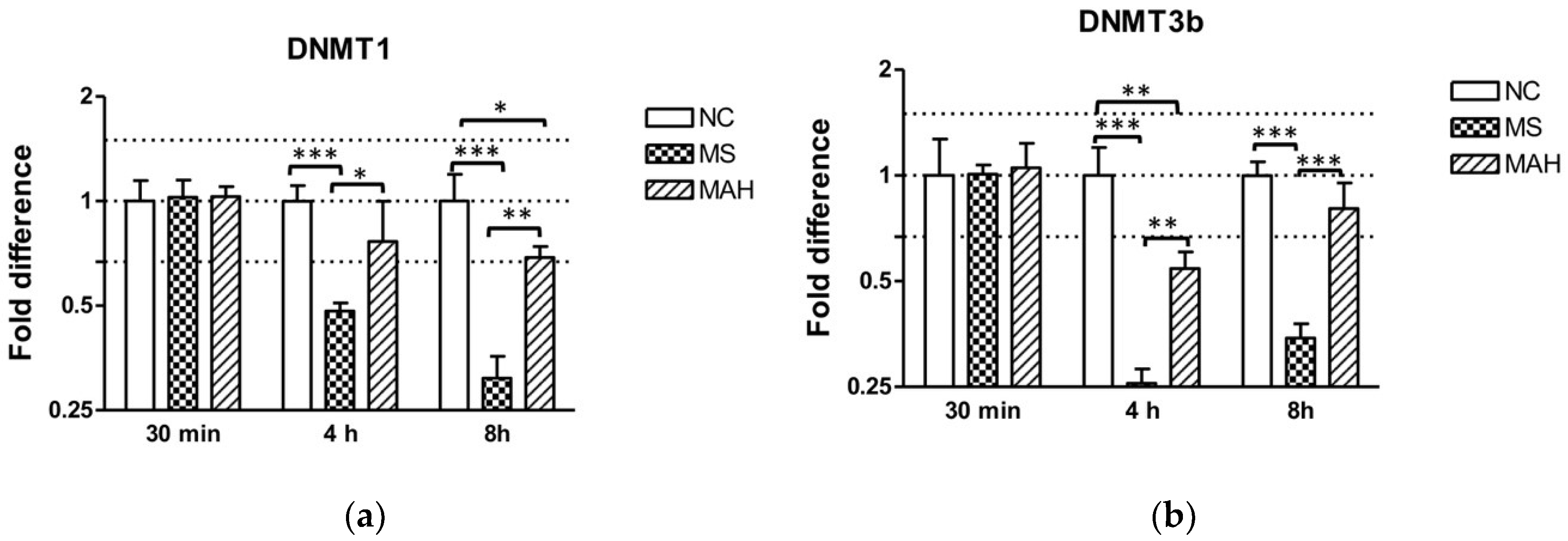
3.2. The Expression of DNA Methlytransferases 1 and 3b Is Downregulated after M. smegmatis Infection
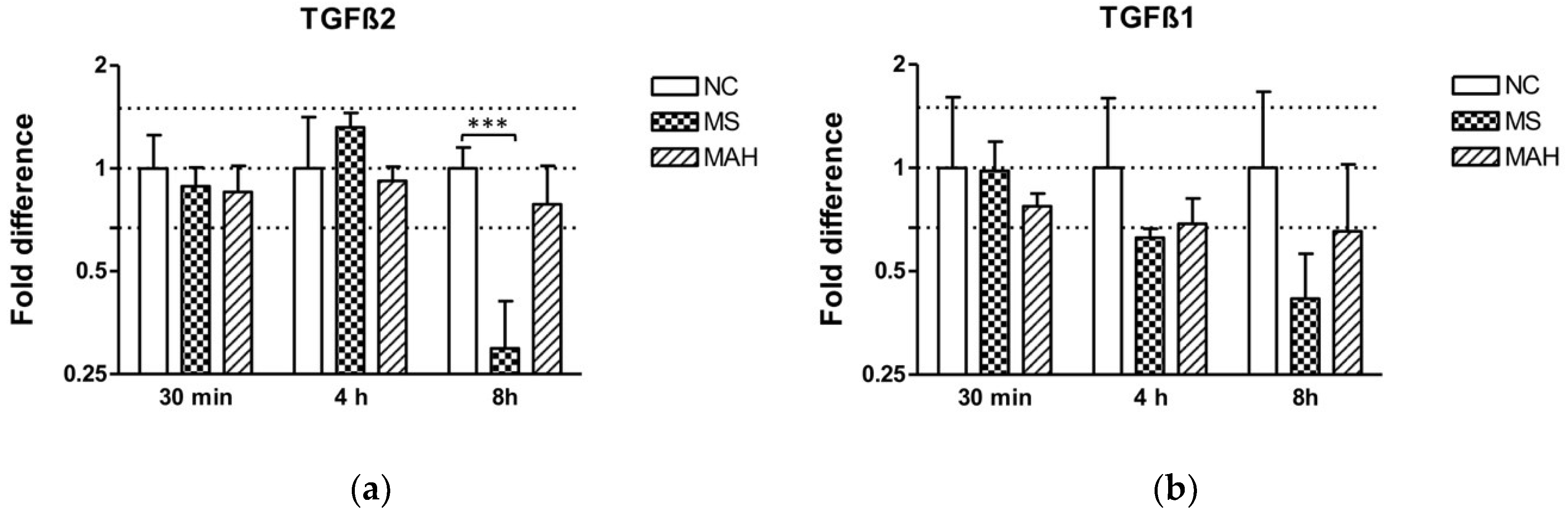
3.3. TGF-β Is Downregulated in M. smegmatis-Infected THP-1-Derived Macrophages
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
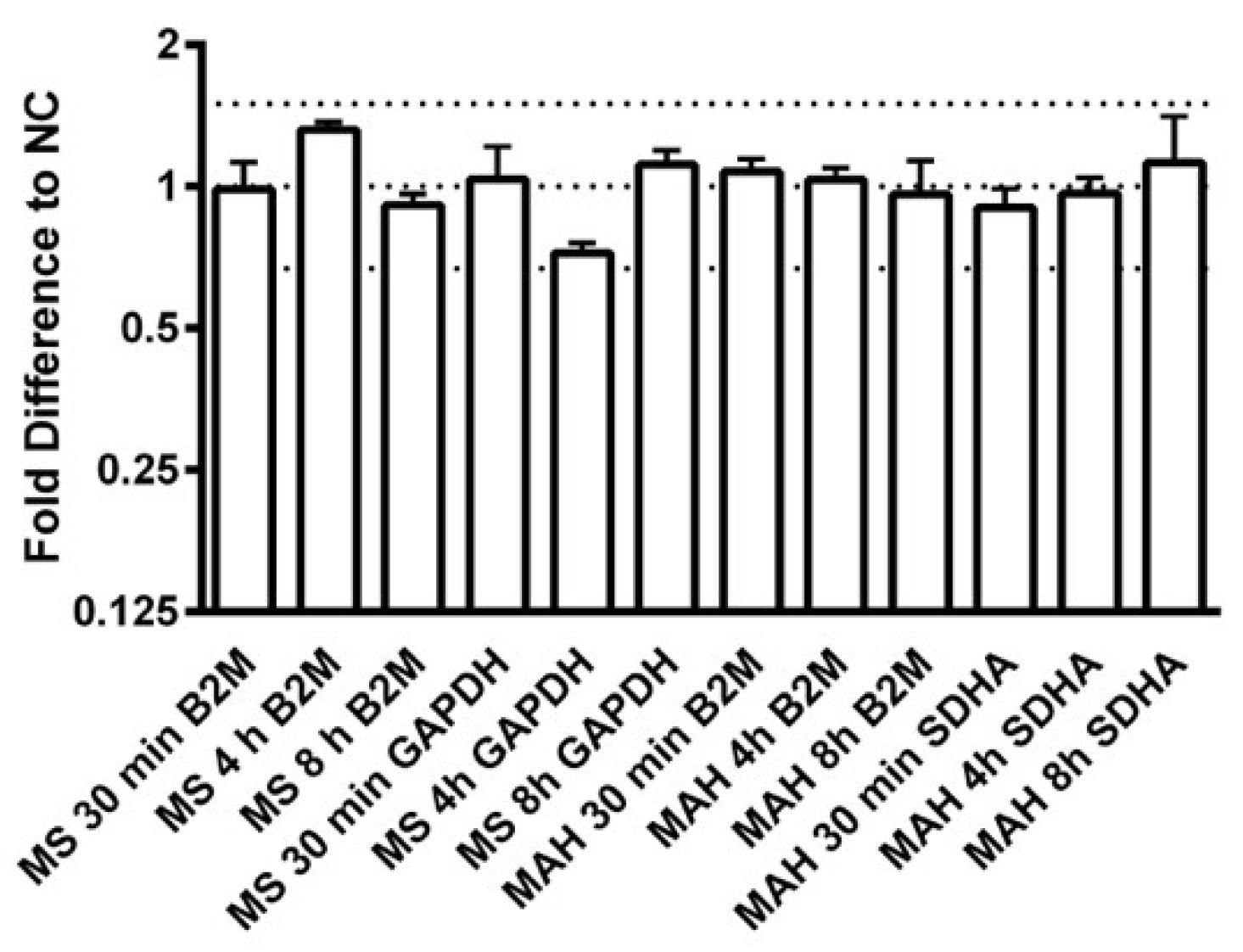
Appendix A
| Gene Name | Forward 5′–3′ | Reverse 5′–3′ | Annealing Temperature |
|---|---|---|---|
| MEG3 | CAGCCAAGGTTCTTGAAAGG | TTCCACGGAGTAGAGGCAGT | 60 °C |
| DNMT1 | GAATCAGTTATGTGACTTGGAAACC | CTAGACGTCCATTCACTTCCC | 60 °C |
| DNMT3b | CCCATTCGAGTCCTGTCATTG | TTGATATTCCCCTCGTGCTTC | 62 °C |
| TGF-β1 | CAGCAACAATTCCTGGCGATA | AAGGCGAAAGCCCTCAATTT | 60 °C |
| TGF-β2 | CCCCGGAGGTGATTTCCATC | CAACTGGGCAGACAGTTTCG | 60 °C |
| B2M | GTGCTCGCGCTACTCTCTCT | GGATGGATGAAACCCAGACA | 60 °C |
| GAPDH | CCATCTTCCAGGAGCGAGAT | CTAAGCAGTTGGTGGTGCAG | 60 °C |
| SDHA | TGGGAACAAGAGGGCATCTG | CCACCACTGCATCAAATTCATG | 60 °C |
Appendix B
References
- Inderlied, C.B.; Kemper, C.A.; Bermudez, L.E. The Mycobacterium avium complex. Clin. Microbiol. Rev. 1993, 6, 266–310. [Google Scholar] [CrossRef] [PubMed]
- Sharbati-Tehrani, S.; Stephan, J.; Holland, G.; Appel, B.; Niederweis, M.; Lewin, A. Porins limit the intracellular persistence of Mycobacterium smegmatis. Microbiology 2005, 151, 2403–2410. [Google Scholar] [CrossRef] [PubMed]
- Von Both, U.; Berk, M.; Agapow, P.M.; Wright, J.D.; Git, A.; Hamilton, M.S.; Goldgof, G.; Siddiqui, N.; Bellos, E.; Wright, V.J.; et al. Mycobacterium tuberculosis Exploits a Molecular Off Switch of the Immune System for Intracellular Survival. Sci. Rep. 2018, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.; Bandeira, M.; Carvalho, P.A.; Duarte, A.; Jordao, L. Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int. J. Mycobacteriol. 2015, 4, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Kuehnel, M.P.; Goethe, R.; Habermann, A.; Mueller, E.; Rohde, M.; Griffiths, G.; Valentin-Weigand, P. Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: Phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell. Microbiol. 2001, 3, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Pawar, K.; Hanisch, C.; Palma Vera, S.E.; Einspanier, R.; Sharbati, S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci. Rep. 2016, 6, 19416. [Google Scholar] [CrossRef]
- Zur Bruegge, J.; Einspanier, R.; Sharbati, S. A Long Journey Ahead: Long Non-coding RNAs in Bacterial Infections. Front. Cell. Infect. Microbiol. 2017, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, J.; Wang, J.; Wen, Q.; Wang, H.; He, J.; Hu, S.; He, W.; Du, X.; Liu, S.; et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 38963. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, H.; Xie, X.; Chen, C.Y.; Huang, D.; Shen, L.; Zhang, H.; Chen, Z.W.; Zeng, G. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc. Natl. Acad. Sci. USA 2015, 112, E3883–E3892. [Google Scholar] [CrossRef] [PubMed]
- L’Abbate, C.; Cipriano, I.; Perez-Hurtado, E.C.; Leao, S.C.; Carneiro, C.R.; Machado, J., Jr. TGF-beta-mediated sustained ERK1/2 activity promotes the inhibition of intracellular growth of Mycobacterium avium in epithelioid cells surrogates. PLoS ONE 2011, 6, e21465. [Google Scholar] [CrossRef]
- Reed, S.G. TGF-beta in infections and infectious diseases. Microbes Infect. 1999, 1, 1313–1325. [Google Scholar] [CrossRef]
- Warsinske, H.C.; Pienaar, E.; Linderman, J.J.; Mattila, J.T.; Kirschner, D.E. Deletion of TGF-beta1 Increases Bacterial Clearance by Cytotoxic T Cells in a Tuberculosis Granuloma Model. Front. Immunol. 2017, 8, 1843. [Google Scholar] [CrossRef] [PubMed]
- Horan, K.L.; Freeman, R.; Weigel, K.; Semret, M.; Pfaller, S.; Covert, T.C.; van Soolingen, D.; Leao, S.C.; Behr, M.A.; Cangelosi, G.A. Isolation of the genome sequence strain Mycobacterium avium 104 from multiple patients over a 17-year period. J. Clin. Microbiol. 2006, 44, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Sharbati, J.; Lewin, A.; Kutz-Lohroff, B.; Kamal, E.; Einspanier, R.; Sharbati, S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS ONE 2011, 6, e20258. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Kruer, T.L.; Dougherty, S.M.; Reynolds, L.; Long, E.; de Silva, T.; Lockwood, W.W.; Clem, B.F. Expression of the lncRNA Maternally Expressed Gene 3 (MEG3) Contributes to the Control of Lung Cancer Cell Proliferation by the Rb Pathway. PLoS ONE 2016, 11, e0166363. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bian, E.B.; He, X.J.; Ma, C.C.; Zong, G.; Wang, H.L.; Zhao, B. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int. J. Oncol. 2016, 48, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Peter, S.; Jung, M.; Lewin, A.; Hemmrich-Stanisak, G.; Franke, A.; von Kleist, M.; Schutte, C.; Einspanier, R.; Sharbati, S.; et al. Analysis of long non-coding RNA and mRNA expression in bovine macrophages brings up novel aspects of Mycobacterium avium subspecies paratuberculosis infections. Sci. Rep. 2019, 9, 1571. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Li, J.; Gao, K.; Fu, Y. Identifcation of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection. J. Infect. 2014, 69, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Babrak, L.; Danelishvili, L.; Rose, S.J.; Kornberg, T.; Bermudez, L.E. The environment of “Mycobacterium avium subsp. Hominissuis” microaggregates induces synthesis of small proteins associated with efficient infection of respiratory epithelial cells. Infect. Immun. 2015, 83, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Maiz Carro, L.; Barbero Herranz, E.; Nieto Royo, R. Respiratory infections due to nontuberculous mycobacterias. Med. Clin. 2018, 150, 191–197. [Google Scholar] [CrossRef]
- Kang, S.H.; Mun, S.K.; Lee, M.J.; Kim, S.Y.; Choi, H.G.; Byun, J.; Kim, C.H.; Kim, H.R.; Cho, S.Y. Endobronchial Mycobacterium avium Infection in an Immunocompetent Patient. Infect. Chemother. 2013, 45, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Prince, D.S.; Peterson, D.D.; Steiner, R.M.; Gottlieb, J.E.; Scott, R.; Israel, H.L.; Figueroa, W.G.; Fish, J.E. Infection with Mycobacterium avium complex in patients without predisposing conditions. N. Engl. J. Med. 1989, 321, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Song, J.Y.; Park, C.W.; Kee, S.Y.; Choi, W.S.; Kang, E.Y.; Sohn, J.W.; Kim, W.J.; Kim, M.J.; Cheong, H.J. Disseminated Mycobacterium avium complex infection in an immunocompetent pregnant woman. BMC Infect. Dis. 2006, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Nathan, C.F.; Graycar, J.; Derynck, R.; Stuehr, D.J.; Srimal, S. Macrophage deactivating factor and transforming growth factors-beta 1 -beta 2 and -beta 3 inhibit induction of macrophage nitrogen oxide synthesis by IFN-gamma. J. Immunol. 1990, 145, 940–944. [Google Scholar] [PubMed]
- Lee, Y.J.; Han, Y.; Lu, H.T.; Nguyen, V.; Qin, H.; Howe, P.H.; Hocevar, B.A.; Boss, J.M.; Ransohoff, R.M.; Benveniste, E.N. TGF-beta suppresses IFN-gamma induction of class II MHC gene expression by inhibiting class II transactivator messenger RNA expression. J. Immunol. 1997, 158, 2065–2075. [Google Scholar] [PubMed]
- Hirsch, C.S.; Yoneda, T.; Averill, L.; Ellner, J.J.; Toossi, Z. Enhancement of intracellular growth of Mycobacterium tuberculosis in human monocytes by transforming growth factor-beta 1. J. Infect. Dis. 1994, 170, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Toossi, Z.; Gogate, P.; Shiratsuchi, H.; Young, T.; Ellner, J.J. Enhanced production of TGF-beta by blood monocytes from patients with active tuberculosis and presence of TGF-beta in tuberculous granulomatous lung lesions. J. Immunol. 1995, 154, 465–473. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharbati, S.; Ravon, F.; Einspanier, R.; zur Bruegge, J. Mycobacterium smegmatis But Not Mycobacterium avium subsp. hominissuis Causes Increased Expression of the Long Non-Coding RNA MEG3 in THP-1-Derived Human Macrophages and Associated Decrease of TGF-β. Microorganisms 2019, 7, 63. https://doi.org/10.3390/microorganisms7030063
Sharbati S, Ravon F, Einspanier R, zur Bruegge J. Mycobacterium smegmatis But Not Mycobacterium avium subsp. hominissuis Causes Increased Expression of the Long Non-Coding RNA MEG3 in THP-1-Derived Human Macrophages and Associated Decrease of TGF-β. Microorganisms. 2019; 7(3):63. https://doi.org/10.3390/microorganisms7030063
Chicago/Turabian StyleSharbati, Soroush, Faustine Ravon, Ralf Einspanier, and Jennifer zur Bruegge. 2019. "Mycobacterium smegmatis But Not Mycobacterium avium subsp. hominissuis Causes Increased Expression of the Long Non-Coding RNA MEG3 in THP-1-Derived Human Macrophages and Associated Decrease of TGF-β" Microorganisms 7, no. 3: 63. https://doi.org/10.3390/microorganisms7030063
APA StyleSharbati, S., Ravon, F., Einspanier, R., & zur Bruegge, J. (2019). Mycobacterium smegmatis But Not Mycobacterium avium subsp. hominissuis Causes Increased Expression of the Long Non-Coding RNA MEG3 in THP-1-Derived Human Macrophages and Associated Decrease of TGF-β. Microorganisms, 7(3), 63. https://doi.org/10.3390/microorganisms7030063





