Fermented Seeds (“Zgougou”) from Aleppo Pine as a Novel Source of Potentially Probiotic Lactic Acid Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Lactic Acid Bacteria (LAB)
2.2. Molecular Biotyping and Identification of LAB
2.3. Probiotic Traits of Lactic Acid Bacteria
2.3.1. DNase and Hemolysis Tests
2.3.2. Antibiotics Susceptibility
2.3.3. Tolerance to Acid, Salt, Pepsin, Pancreatin, and Bile Salt
2.3.4. Biofilm Production, Adhesion, and Aggregation Capacities
2.3.5. Cell Surface Hydrophobicity Test
2.3.6. Enzyme Activities
2.3.7. Antimicrobial Activity
2.3.8. Inhibition of Biofilm Production
2.3.9. Antioxidant Activity
2.4. Statistical Analyses
3. Results
3.1. Safety of Lactic Acid Bacteria (LAB) Strains Identified from Zgougou
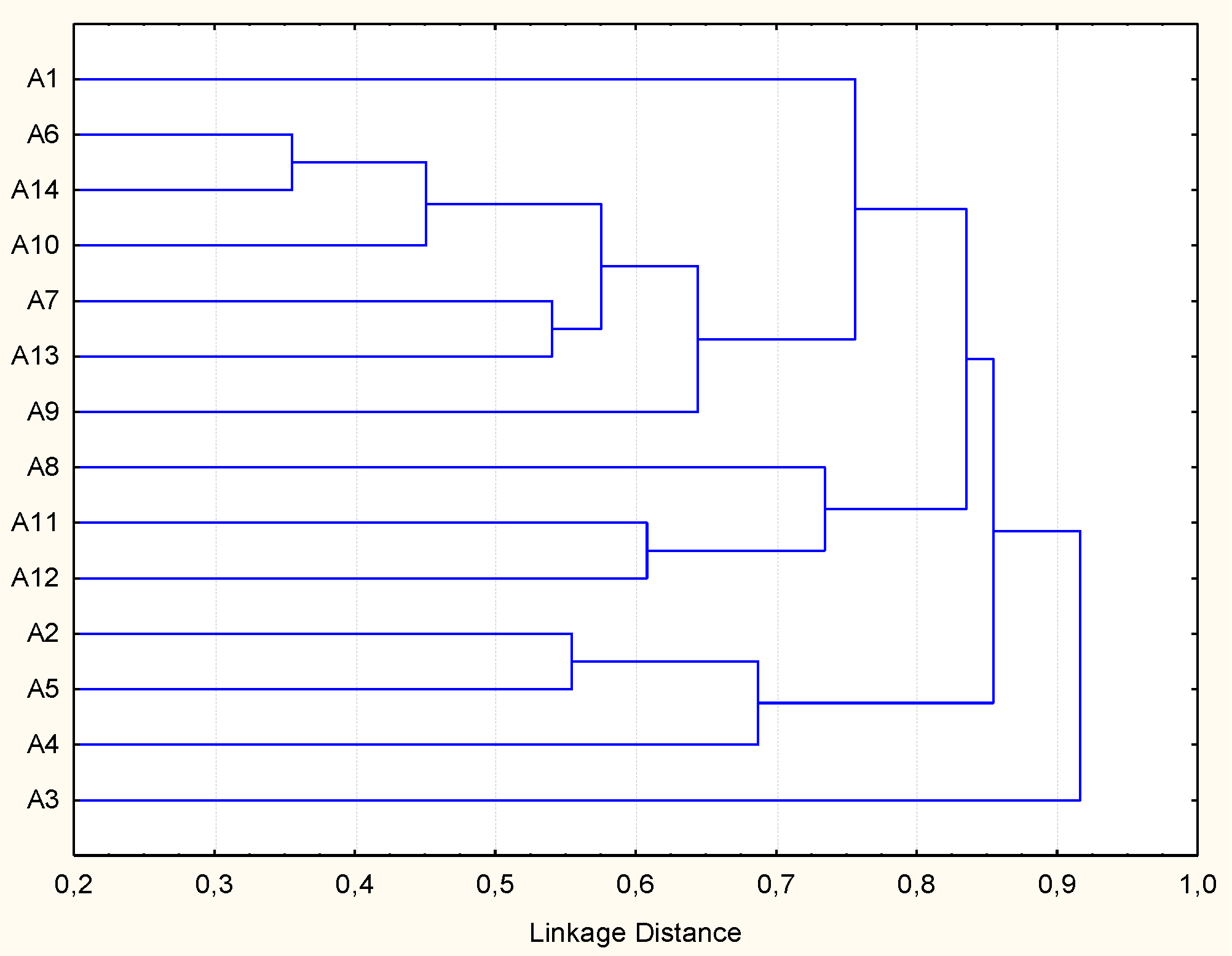
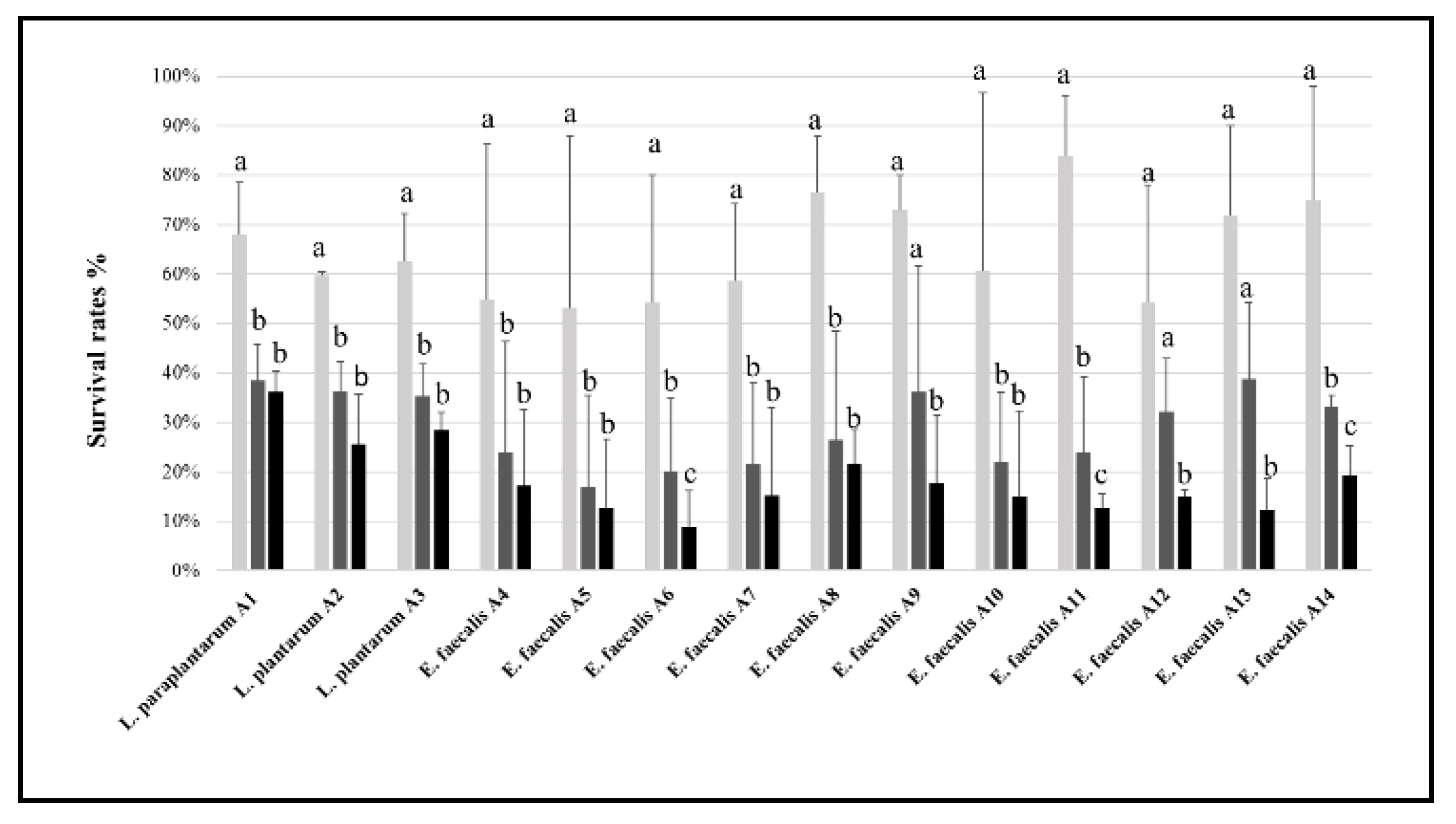
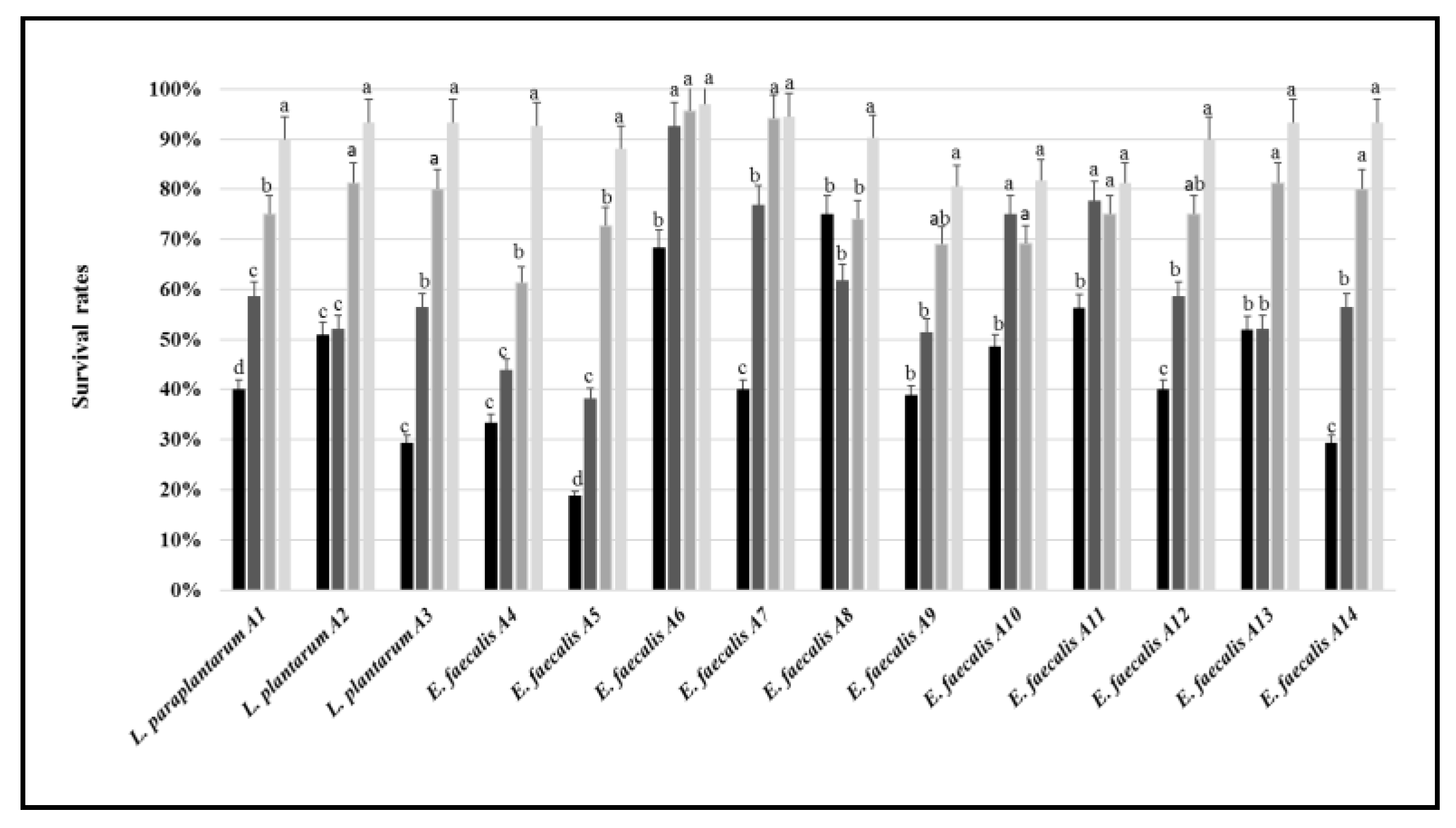
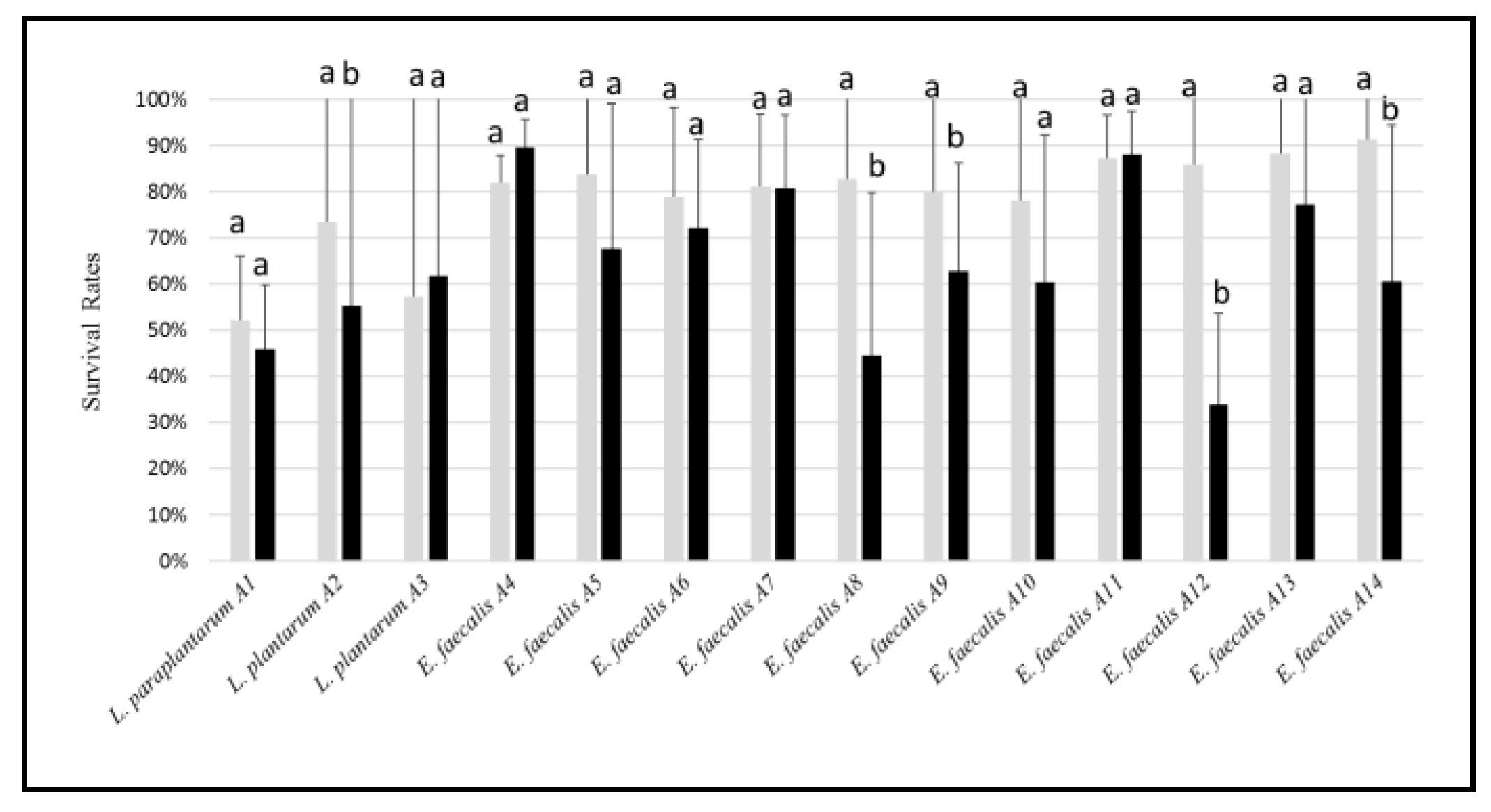
3.2. Tolerance to Stressing Environmental Conditions
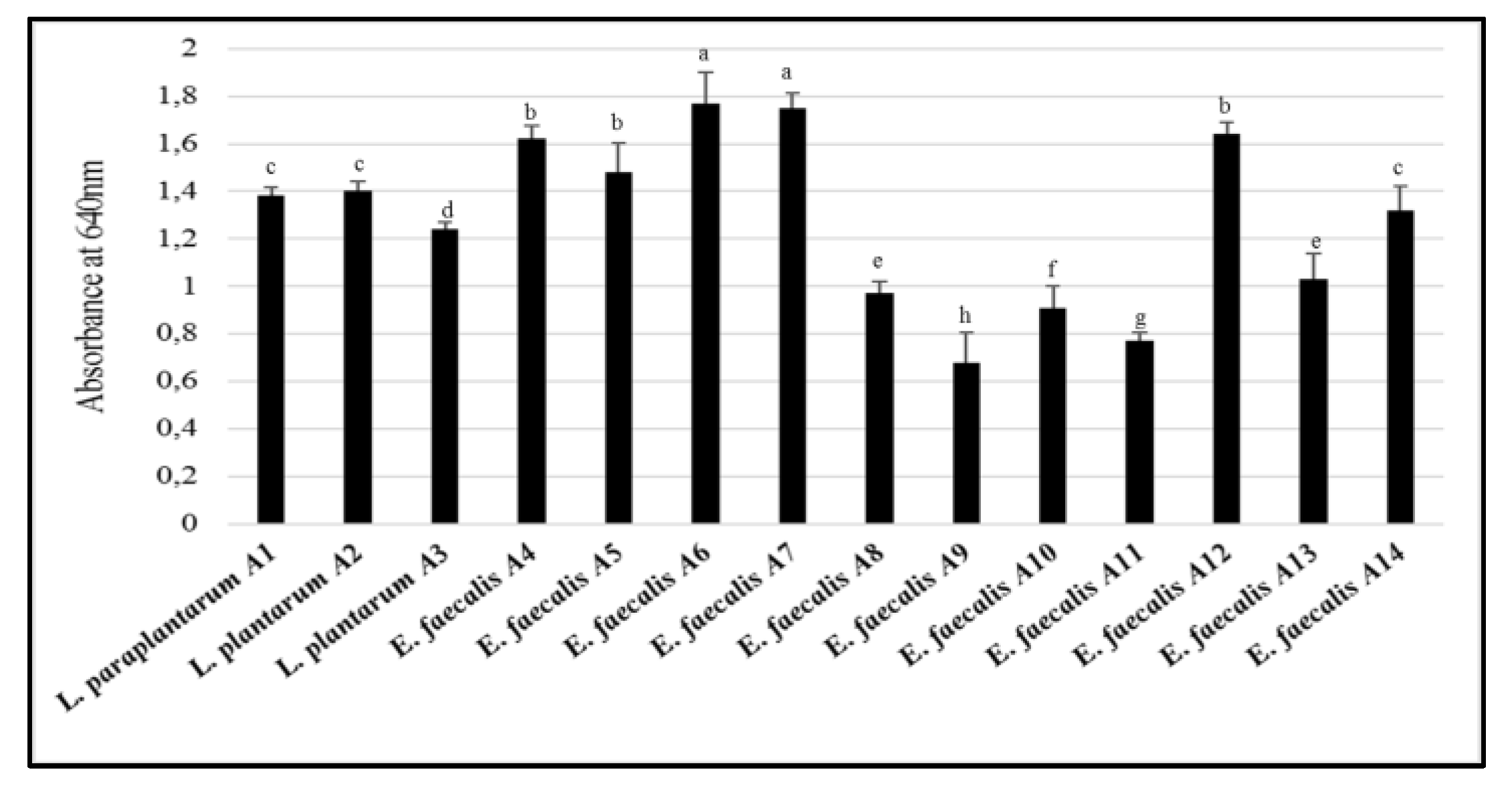
3.3. Biofilm Production, Adhesion and Aggregation Capacities
3.4. Cell Surface Hydrophobicity Test
3.5. Enzyme Activities
3.6. Antibacterial Activity
3.7. Antifungal Activity
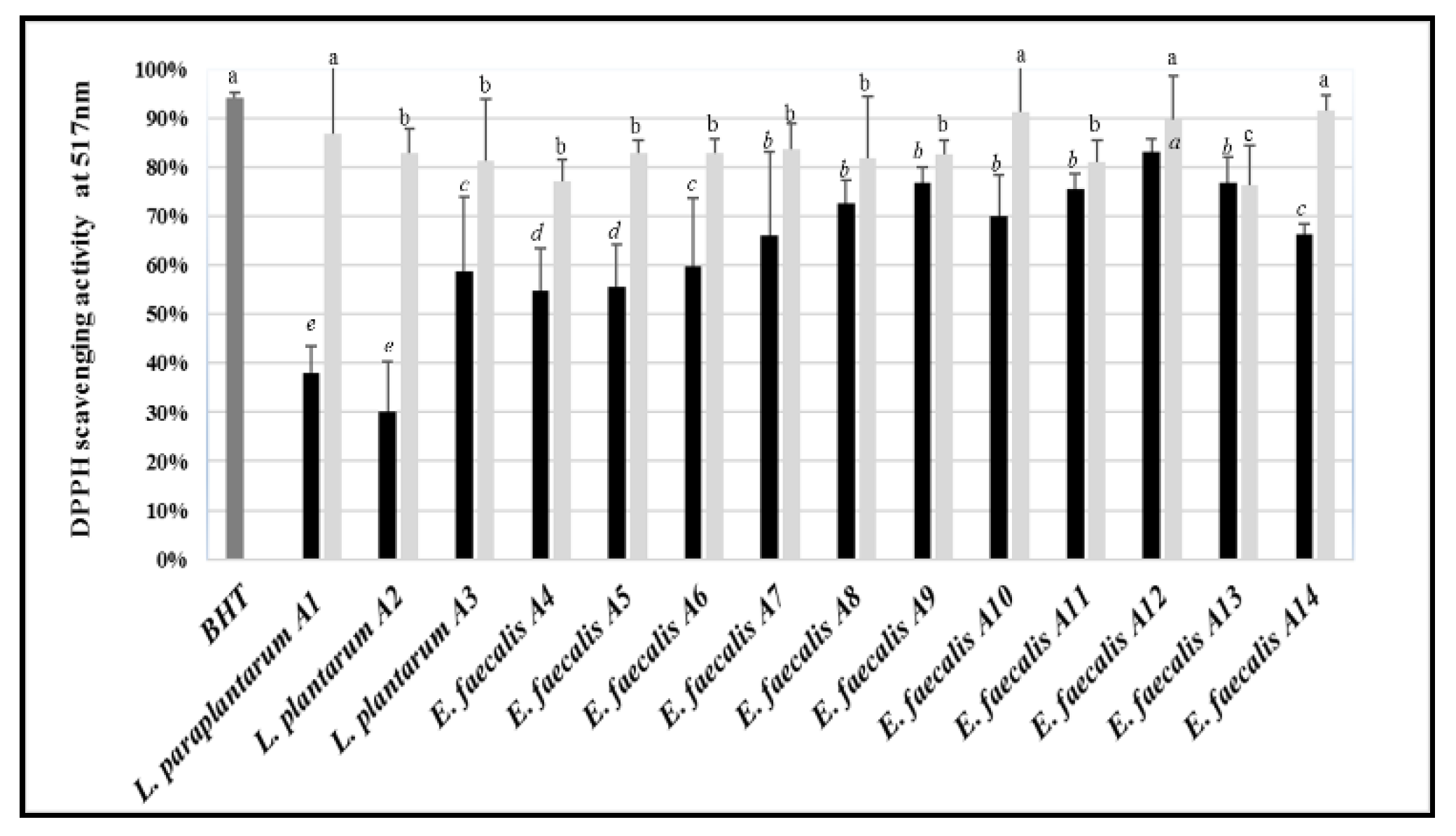
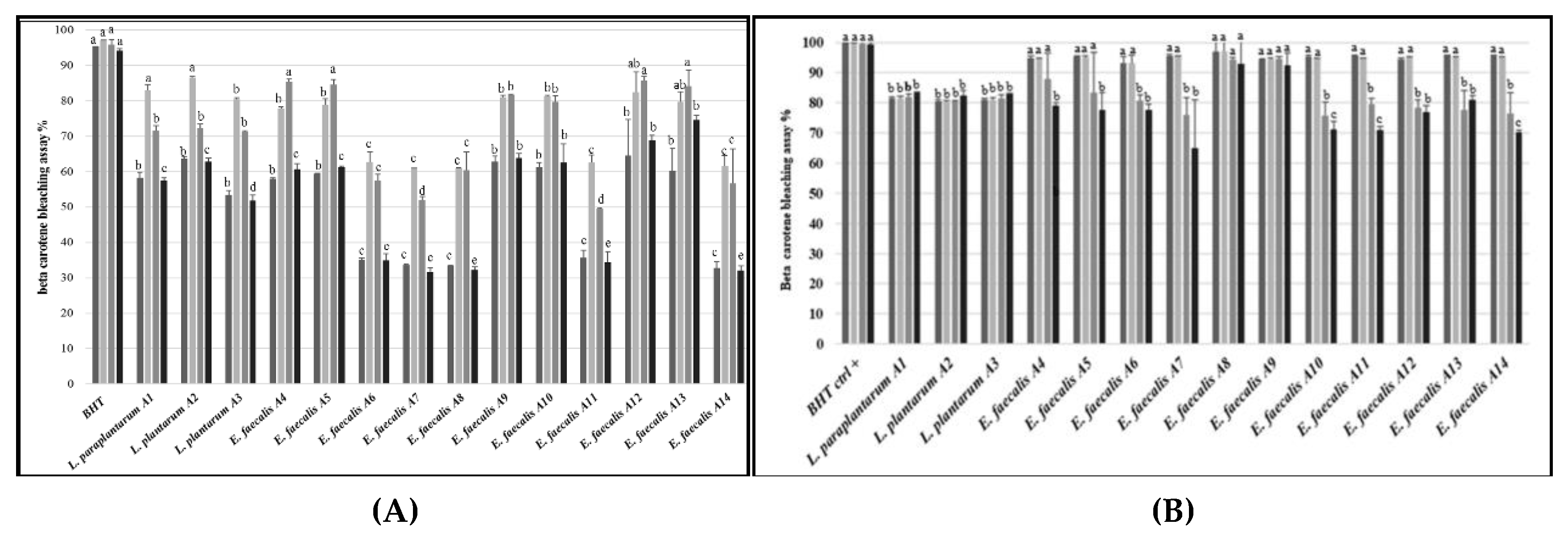
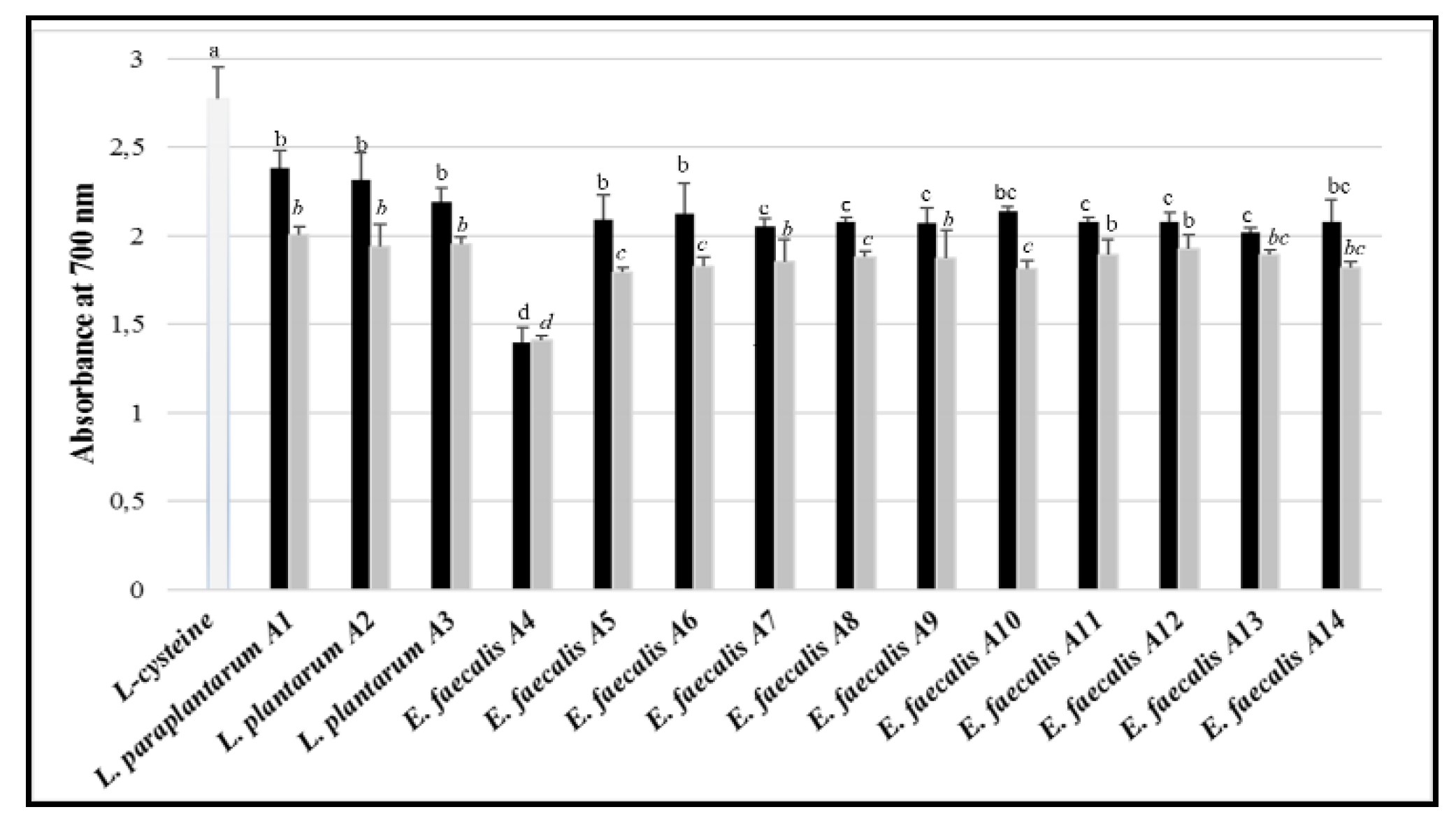
3.8. Antioxidant Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Soccol, C.R.; Vandenberghe, L.P.D.; Spier, M.R.; Medeiros, A.B.P.; Yamaguishi, C.T.; Lindner, J.D.; Pandey, A.; Thomaz-Soccol, V. The potential of probiotics: A review. Food Technol. Biotechnol. 2010, 48, 413–434. [Google Scholar]
- Oelschlaeger, T.A. Mechanisms of probiotic actions–A review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Abrunhosa, L.; Inês, A.; Rodrigues, A.I.; Guimarães, A.; Pereira, V.L.; Parpot, P.; Mendes-Faia, A.; Venâncio, A. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Med. Microbiol. 2014, 188, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Le Lay, C.; Mounier, J.; Vasseur, V.; Weill, A.; Le Blay, G.; Barbier, G.; Coton, E. In vitro and in situ screening of lactic acid bacteria and propionibacteria antifungal activities against bakery product spoilage molds. Food Control 2016, 60, 247–255. [Google Scholar] [CrossRef]
- Araújo, T.F.; Ferreira, C.L.d.L.F. The genus Enterococcus as probiotic: Safety concerns. Braz. Arch. Biol. Technol. 2013, 56, 457–466. [Google Scholar] [CrossRef]
- Shetty, P.H.; Jespersen, L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 2006, 17, 48–55. [Google Scholar] [CrossRef]
- Rima, H.; Ismail, F. Antimicrobial and probiotic properties of yeasts: From fundamental to novel applications. Front. Microbiol. 2012, 3, 421. [Google Scholar]
- O’Shea, E.F.; Cotter, P.D.; Stanton, C.; Ross, R.P.; Hill, C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: Bacteriocins and conjugated linoleic acid. Int. J. Food Microbiol. 2012, 152, 189–205. [Google Scholar] [CrossRef]
- Rathore, M.; Sharma, K. Probiotics and their Indian and global value: A review. World J. Pharm. Med. Res. 2017, 3, 43–54. [Google Scholar]
- Van den Berg, D.J.; Smits, A.; Potn, B.; Ledeboer, A.M.; Kersters, K.; Verbake, J.M.; Verrips, C.T. Isolation, screening and identification of lactic acid bacteria from traditional food fermentation processes and culture collections. Food Biotechnol. 1993, 7, 189–205. [Google Scholar] [CrossRef]
- Ali, A.A.; Mustafa, M.M. Isolation, characterization and identification of lactic acid bacteria from fermented sorghum dough used in Sudanese Kisra preparation. Pak. J. Nutr. 2009, 8, 1814–1818. [Google Scholar] [CrossRef][Green Version]
- Manini, F.; Casiraghi, M.C.; Poutanen, K.; Brasca, M.; Erba, D.; Plumed-Ferrer, C. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT Food Sci. Technol. 2016, 66, 275–283. [Google Scholar] [CrossRef]
- Alfonzo, A.; Ventimiglia, G.; Corona, O.; Di Gerlando, R.; Gaglio, R.; Francesca, N.; Moschetti, G.; Settanni, L. Diversity and technological potential of lactic acid bacteria of wheat flours. Food Microbiol. 2013, 36, 343–354. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Júnior, W.L.G.; Da Silva Ferrari, Í.; De Souza, J.V.; Da Silva, C.D.A.; Da Costa, M.M.; Dias, F.S. Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 2015, 53, 96–103. [Google Scholar] [CrossRef]
- Wang, D.; Liu, W.; Ren, Y.; De, L.; Zhang, D.; Yang, Y.; Bao, Q.; Zhang, H.; Menghe, B. Isolation and Identification of lactic acid bacteria from traditional dairy products in Baotou and Bayannur of midwestern inner Mongolia and q-PCR analysis of predominant species. Korean J. Food Sci. An. 2016, 36, 499. [Google Scholar] [CrossRef]
- Verón, H.E.; Di Risio, H.D.; Isla, M.I.; Torres, S. Isolation and selection of potential probiotic lactic acid bacteria from Opuntia ficus-indica fruits that grow in Northwest Argentina. LWT Food Sci. Technol. 2017, 84, 231–240. [Google Scholar] [CrossRef]
- Naeem, M.; Ilyas, M.; Haider, S.; Baig, S.; Saleem, M. Isolation characterization and identification of lactic acid bacteria from fruit juices and their efficacy against antibiotics. Pak. J. Bot. 2012, 44, 8. [Google Scholar]
- Tajabadi, N.; Mardan, M.; Manap, M.Y.A.; Mustafa, S. Molecular identification of Lactobacillus spp. isolated from the honey comb of the honey bee (Apis dorsata) by 16S rRNA gene sequencing. J. Apic. Res. 2013, 52, 235–241. [Google Scholar] [CrossRef]
- Ekundayo, F. Isolation and identification of lactic acid bacteria from rhizosphere soils of three fruit trees, fish and ogi. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 991–998. [Google Scholar]
- Kaur, R.; Tiwari, S. Isolation, identification and characterization of Pediococcus pentosaceus LB44 and Weissella confusa LM85 for the presence of bacteriocin-like inhibitory substances (BLIS). Microbiology 2016, 85, 540–547. [Google Scholar] [CrossRef]
- Fuchs-Tarlovsky, V.; Marquez-Barba, M.F.; Sriram, K. Probiotics in dermatologic practice. Nutr. J. 2016, 32, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, A.D.; Paz, M.L.; Leoni, J.; Gonzalez Maglio, D.H. Message in a bottle: Dialog between intestine and skin modulated by probiotics. Int. J. Mol. Sci. 2017, 18, 1067. [Google Scholar] [CrossRef] [PubMed]
- Tsitsoni, T.K. Seed quality characteristics of Pinus halepensis–seed germination strategy and early seedling growth. Web Ecol. 2009, 9, 72–76. [Google Scholar] [CrossRef]
- Fekih, N.; Allali, H.; Merghache, S.; Chaïb, F.; Merghache, D.; El Amine, M.; Djabou, N.; Muselli, A.; Tabti, B.; Costa, J. Chemical composition and antibacterial activity of Pinus halepensis Miller growing in West Northern of Algeria. Asian Pac. J. Trop. Dis. 2014, 4, 97. [Google Scholar] [CrossRef]
- Nasri, N.; Triki, S. Analyse des lipides des graines de pins de Tunisie (Pinus halepensis Mill. et Pinus pinea L.). Riv. Ital. Sostanze Grasse 2004, 81, 244–247. [Google Scholar]
- Cheikh-Rouhou, S.; Hentati, B.; Besbes, S.; Blecker, C.; Deroanne, C.; Attia, H. Chemical composition and lipid fraction characteristics of Aleppo pine (Pinus halepensis Mill.) seeds cultivated in Tunisia. Food Sci. Technol. Int. 2006, 12, 407–415. [Google Scholar] [CrossRef]
- Nahal, I. Taxonomie et aire géographique des pins du groupe halepensis. CIHEAM Options Méditerranéennes 1986, 1, 1–9. [Google Scholar]
- Stendid, J.; Karlsson, J.O.; Hogberg, N. Intra-specific genetic variation in Heterobasidium annosum revealed by amplification of minisatellite DNA. Mycol. Res. 1994, 98, 57–63. [Google Scholar] [CrossRef]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- Zapparoli, G.; Torriani, S.; and Dell’aglio, F. Differentiation of Lactobacillus sanfranciscensis strains by randomly amplified polymorphic DNA and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 1998, 166, 324–332. [Google Scholar] [CrossRef][Green Version]
- De Angelis, M.; Siragusa, S.; Berloco, M.; Caputo, L.; Settanni, L.; Alfonsi, G.; Amerio, M.; Grandi, A.; Ragni, A.; Gobbetti, M. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 2006, 157, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Rizzello, C.G.; Gagliardi, F.; Ricciuti, P.; Ndagijimana, M.; Francavilla, R.; Guerzoni, M.E.; Crecchio, C.; Gobbetti, M.; De Angelis, M. Different fecal microbiotas and volatile organic compounds in treated and untreated children with celiac disease. Appl. Environ. Microbiol. 2009, 75, 3963–3971. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Aguas, R.; Ferguson, N.M. Feature selection methods for identifying genetic determinants of host species in RNA viruses. PLoS Comput. Biol. 2013, 9, e1003254. [Google Scholar]
- Thapa, N.; Pal, J.; Tamang, J.P. Microbial diversity in ngari, hentak and tungtap, fermented fish products of North-East India. World J. Microbiol. Biotechnol. 2004, 20, 599. [Google Scholar] [CrossRef]
- Vesterlund, S.; Paltta, J.; Karp, M.; Ouwehand, A.C. Adhesion of bacteria to resected human colonic tissue: Quantitative analysis of bacterial adhesion and viability. Res. Microbiol. 2005, 156, 238–244. [Google Scholar] [CrossRef]
- Taheur, F.B.; Kouidhi, B.; Fdhila, K.; Elabed, H.; Slama, R.B.; Mahdouani, K.; Bakhrouf, A.; Chaieb, K. Anti-bacterial and anti-biofilm activity of probiotic bacteria against oral pathogens. Microb. Pathog. 2016, 97, 213–220. [Google Scholar] [CrossRef]
- Ambalam, P.; Kondepudi, K.K.; Nilsson, I.; Wadström, T.; Ljungh, Å. Bile stimulates cell surface hydrophobicity, Congo red binding and biofilm formation of Lactobacillus strains. FEMS Microbiol. Lett. 2012, 333, 10–19. [Google Scholar]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef]
- Maria, G.; Sridhar, K.; Raviraja, N. Antimicrobial and enzyme activity of mangrove endophytic fungi of southwest coast of India. J. Agric. Technol. 2005, 1, 67–80. [Google Scholar]
- Carrim, A.J.I.; Barbosa, E.C.; Vieira, J.D.G. Enzymatic activity of endophytic bacterial isolates of Jacaranda decurrens Cham. (Carobinha-do-campo). Braz. Arch. Biol. Technol. 2006, 49, 353–359. [Google Scholar] [CrossRef]
- Hashemi, S.M.B.; Mousavi-Khaneghah, A.; Kontominas, M.G.; Eş, I.; Sant’Ana, A.S.; Martinez, R.R.; Drider, D. Fermentation of sarshir (kaymak) by lactic acid bacteria: Antibacterial activity, antioxidant properties, lipid and protein oxidation and fatty acid profile. J. Sci. Food Agric. 2017, 97, 4595–4603. [Google Scholar] [CrossRef] [PubMed]
- Cherif, A.; Sadfi-Zouaoui, N.; Eleuch, D.; Dhahri, A.B.O.; Boudabous, A. Pseudomonas isolates have in vitro antagonistic activity against the dermatophytes Trichophyton rubrum, Trichophyton mentagrophytes var interdigitale and Microsporum canis. J. Mycol. Med. 2009, 19, 178–184. [Google Scholar] [CrossRef]
- Whipps, J.M. Effect of media on growth and interactions between a range of soil-borne glasshouse pathogens and antagonistic fungi. New Phytol. 1987, 107, 127–142. [Google Scholar] [CrossRef]
- Korsten, L.; De Jager, E.; De Villiers, E.; Lourens, A.; Kotze, J.; Wehner, F. Evaluation of bacterial epiphytes isolated from avocado leaf and fruit surfaces. Plant Dis. 1997, 9, 1149. [Google Scholar]
- Wang, H.; Yan, Y.; Wang, J.; Zhang, H.; Qi, W. Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PLoS ONE 2012, 7, e29452. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disc Susceptibility Tests, 13th ed.; CLSI standard M02; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Bougatef, A.; Nedjar-Arroume, N.; Manni, L.; Ravallec, R.; Barkia, A.; Guillochon, D.; Nasri, M. Purification and identification of novel antioxidant peptides from enzymatic hydrolysates of sardinelle (Sardinella aurita) by-products proteins. Food Chem. 2010, 118, 559–565. [Google Scholar] [CrossRef]
- Trabelsi, I.; Ktari, N.; Slima, S.B.; Triki, M.; Bardaa, S.; Mnif, H.; Salah, R.B. Evaluation of dermal wound healing activity and in vitro antibacterial and antioxidant activities of a new exopolysaccharide produced by Lactobacillus sp. Ca6. Int. J. Biol. Macromol. 2017, 103, 194–201. [Google Scholar] [CrossRef]
- Son, S.-H.; Yang, S.-J.; Jeon, H.-L.; Yu, H.-S.; Lee, N.-K.; Park, Y.-S.; Park, H.-D. Antioxidant and immunostimulatory effect of potential probiotic Lactobacillus paraplantarum SC61 isolated from Korean traditional fermented food, jangajji. Microb. Pathog. 2018, 125, 486–492. [Google Scholar] [CrossRef]
- Tamang, J.P.; Shin, D.-H.; Jung, S.-J.; Chae, S.-W. Functional properties of microorganisms in fermented foods. Front. Microbiol. 2016, 7, 578. [Google Scholar] [CrossRef]
- Chilton, S.N.; Burton, J.P.; Reid, G. Inclusion of fermented foods in food guides around the world. Nutrients 2015, 7, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Metchnikoff, E. The Prolongation of Life: Optimistic Studies; G.P. Putnam’s Sons: New York, NY, USA, 1908. [Google Scholar]
- Derrien, M.; van Hylckama Vlieg, J.E. Fate activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015, 520, 104. [Google Scholar] [CrossRef]
- Que, S.K.T.; Zwald, F.O.; Schmults, C.D. Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J. Am. Acad. Dermatol. 2018, 78, 237–247. [Google Scholar] [CrossRef]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Microbes Infect. 2002, 4, 481–489. [Google Scholar] [CrossRef]
- Nole, K.L.B.; Yim, E.; Keri, J.E. Probiotics and prebiotics in dermatology. J. Am. Acad. Dermatol. 2014, 71, 814–821. [Google Scholar] [CrossRef]
- Del Papa, M.F.; Hancock, L.E.; Thomas, V.C.; Perego, M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacterial. 2007, 189, 8835–8843. [Google Scholar] [CrossRef]
- Nishida, S.; Ishii, M.; Nishiyama, Y.; Abe, S.; Ono, Y.; Sekimizu, K. Lactobacillus paraplantarum 11-1 isolated from rice bran pickles activated innate immunity and improved survival in a silkworm bacterial infection model. Front. Microbiol. 2017, 8, 436. [Google Scholar] [CrossRef]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef]
- Annette, C.A.; Daniel, J.; Ingrid, H.; Lamprini, K.; Johan, W.; Elmar, H.; Nicole, A.; Kirstin, V.; Annette, W.; Al-Ahmad, A. Enterococcus faecalis from food, clinical specimens, and oral sites: Prevalence of virulence factors in association with biofilm formation. Front. Microbiol. 2016, 6, 1534. [Google Scholar]
- Koel, B.; Puja, S.; Joshi, S.R. Co-occurrence of antimicrobial resistance and virulence determinants in enterococci isolated from traditionally fermented fish products. J. Glob. Antimicrob. Resist. 2019, 17, 79–83. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Opinion of the scientific panel on additives and products or substances used in animal feed on the updating of the criteria used in assessment of bacteria for resistance to antibiotics of human and veterinary importance. EFSA J. 2005, 223, 1–12. [Google Scholar]
- Ammor, M.S.; Mayo, B. Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: An update. Meat Sci. 2007, 76, 138–146. [Google Scholar] [CrossRef]
- Endo, A.; Sasaki, F.; Maeno, S.; Kanesaki, Y.; Hamaguchi, Y.; Torres, G.A.; Tomita, S.; Nakagawa, J. In vitro and in silico characterisation of Lactobacillus paraplantarum D2-1, a starter culture for soymilk fermentation. Int. J. Food Sci. Nutr. 2018, 69, 857–869. [Google Scholar] [CrossRef]
- Ahn, C.; Collins-Thompson, D.; Duncan, C.; Stiles, M.E. Mobilization and location of the genetic determinant of chloramphenicol resistance from Lactobacillus plantarum caTC2R. Plasmid 1992, 27, 169–176. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef]
- Roberts, M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef]
- de Fátima Silva Lopesa, M.; Ribeiro, T.; Abrantes, M.; Figueiredo Marques, J.J.; Tenreiro, R.; Barreto Crespo, M.T. Antimicrobial resistance profiles of dairy and clinical isolates and type strains of enterococci. Int. J. Food Microbiol. 2005, 103, 191–198. [Google Scholar] [CrossRef]
- Nueno-Palop, C.; Narbad, A. Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 2011, 145, 390–394. [Google Scholar] [CrossRef]
- Muñoz, M.dC.C.; Benomar, N.; Lerma, L.L.; Gálvez, A.; Abriouel, H. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 2014, 172, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.F.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Eliane, E.; Patricia Jeanne, dS.M.M.; João, F.G.; Evanovich, E.; de Souza Mendonça Mattos, P.J.; Farias Guerreiro, J. Comparative genomic analysis of Lactobacillus plantarum: An overview. Int. J. Genom. 2019, 2019, 4973214. [Google Scholar]
- FAO/WHO. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO/WHO: London, ON, Canada, 2002. [Google Scholar]
- Oh, Y.J.; Jung, D.S. Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT Food Sci. Technol. 2015, 63, 437–444. [Google Scholar] [CrossRef]
- Foulquié, M.M.; Callewaert, R.; Devreese, B.; Van Beeumen, J.; De Vuyst, L. Isolation and biochemical characterisation of enterocins produced by enterococci from different sources. J. Appl. Microbiol. 2003, 94, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Tokatlı, M.; Gülgör, G.; Bağder Elmacı, S.; Arslankoz İşleyen, N.; Özçelik, F. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. BioMed Res. Int. 2015, 315819. [Google Scholar]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef]
- Mishra, A.K.; Ghosh, A.R. Characterization of functional, safety, and probiotic properties of Enterococcus faecalis AG5 isolated from wistar rat, demonstrating adherence to HCT 116 cells and gastrointestinal survivability. Probiotics Antimicrob. Proteins 2018, 10, 435–445. [Google Scholar] [CrossRef]
- Cui, X.; Shi, Y.; Gu, S.; Yan, X.; Chen, H.; Ge, J. Antibacterial and antibiofilm activity of lactic acid bacteria isolated from traditional artisanal milk cheese from northeast China against enteropathogenic bacteria. Probiotics Antimicrob. Proteins 2017, 10, 601–610. [Google Scholar] [CrossRef]
- Lleò, M.dM.; Bonato, B.; Benedetti, D.; Canepari, P. Survival of enterococcal species in aquatic environments. FEMS Microbiol. Ecol. 2005, 54, 189–196. [Google Scholar] [CrossRef]
- Liu, L.; Wu, R.; Zhang, J.; Li, P. Overexpression of LuxS promotes stress resistance and biofilm formation of Lactobacillus paraplantarum L-ZS9 by regulating the expression of multiple genes. Front. Microbiol. 2018, 9, e02628. [Google Scholar] [CrossRef] [PubMed]
- Meira, S.M.M.; Helfer, V.E.; Velho, R.V.; Lopes, F.C.; Brandelli, A. Probiotic potential of Lactobacillus spp. isolated from Brazilian regional ovine cheese. J. Dairy Res. 2012, 79, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, N.; Yamazaki, S. Probiotics and safety. Am. J. Clin. Nutr. 2001, 73, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Lauková, A.; Kandričáková, A.; Buňková, L.; Pleva, P.; Ščerbová, J. Sensitivity to enterocins of biogenic amine-producing faecal enterococci from ostriches and pheasants. Probiotics Antimicrob. Proteins 2017, 9, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Van Zyl, W.; Deane, S.; Dicks, L. Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 excludes Listeria monocytogenes from the GIT, as shown by bioluminescent studies in mice. Benefic. Microbes 2016, 7, 227–235. [Google Scholar] [CrossRef]
- Schär-Zammaretti, P.; Ubbink, J. The cell wall of lactic acid bacteria: Surface constituents and macromolecular conformations. Biophys. J. 2003, 85, 4076–4092. [Google Scholar] [CrossRef]
- Osmanagaoglu, O.; Kiran, F.; Ataoglu, H. Evaluation of in vitro probiotic potential of Pediococcus pentosaceus OZF isolated from human breast milk. Probiotics Antimicrob. Proteins 2010, 2, 162–174. [Google Scholar] [CrossRef]
- Devi, S.M.; Aishwarya, S.; Halami, P.M. Discrimination and divergence among Lactobacillus plantarum-group (LPG) isolates with reference to their probiotic functionalities from vegetable origin. Syst. Appl. Microbiol. 2016, 39, 562–570. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Hyrslova, I.; Krausova, G.; Bartova, J.; Kolesar, L.; Jaglic, Z.; Stankova, B.; Curda, L. Characterization of Enterococcus faecium CCDM 922 in respect of its technological and probiotic properties. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 474–482. [Google Scholar] [CrossRef]
- De Vuyst, L.; Moreno, M.F.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Thakkar, P.; Modi, H.; Prajapati, J. Isolation, characterization and safety assessment of lactic acid bacterial isolates from fermented food products. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 713–725. [Google Scholar]
- Meyers, S.; Cuppett, S.; Hutkins, R. Lipase production by lactic acid bacteria and activity on butter oil. Food Microbiol. 1996, 13, 383–389. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: Between probiotic potential and safety concerns—An update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 2004, 70, 5715–5731. [Google Scholar] [CrossRef]
- Askarian, F.; Kousha, A.; Salma, W.; Ringø, E. The effect of lactic acid bacteria administration on growth, digestive enzyme activity and gut microbiota in Persian sturgeon (Acipenser persicus) and beluga (Huso huso) fry. Aquacult. Nutr. 2011, 17, 488–497. [Google Scholar] [CrossRef]
- Fossi, B.T.; Tavea, F. Application of Amylolytic Lactobacillus Fermentum 04BBA19 in Fermentation for Simultaneous Production of Thermostable Alpha-Amylase and Lactic Acid. In Lactic Acid Bacteria-R & D for Food, Health and Livestock Purposes; Kongo, M., Ed.; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef][Green Version]
- Padmavathi, T.; Bhargavi, R.; Priyanka, P.R.; Niranjan, N.R.; Pavitra, P.V. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J Genet. Eng. Biotechnol. 2018, 16, 357–362. [Google Scholar] [CrossRef]
- Kalui, C.; Mathara, J.; Kutima, P.; Kiiyukia, C.; Wongo, L. Functional characteristics of Lactobacillus plantarum and Lactobacillus rhamnosus from ikii, a Kenyan traditional fermented maize porridge. Afr. J. Biotechnol. 2009, 8, 4363–4373. [Google Scholar]
- Kumar, B.B.; Claes, I.J.; Lebeer, S. Functional mechanisms of probiotics. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 321–327. [Google Scholar]
- Ouwehand, A.C.; Lahtinen, S.; Tiihonen, K. The potential of probiotics and prebiotics for skin health. In Textbook of Aging Skin; Farage, M., Miller, K., Maibach, H., Eds.; Springer: Berlin, Germany, 2015; pp. 1299–1313. [Google Scholar]
- O’Neill, C.A.; Monteleone, G.; McLaughlin, J.T.; Paus, R. The gut-skin axis in health and disease: A paradigm with therapeutic implications. BioEssays 2016, 38, 1167–1176. [Google Scholar] [CrossRef]
- Seite, S.; Bieber, T. Barrier function and microbiotic dysbiosis in atopic dermatitis. Clin. Cosmet. Investig. Dermatol. 2015, 8, 479. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.L.V.; Leite, A.Z.; Higuchi, B.S.; Gonzaga, M.I.; Mariano, V.S. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 2017, 152, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; de Melo Franco, B.D. Use of potential probiotic lactic acid bacteria (LAB) biofilms for the control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157: H7 biofilms formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed]
- Belgacem, Z.B.; Abriouel, H.; Omar, N.B.; Lucas, R.; Martínez-Canamero, M.; Gálvez, A.; Manai, M. Antimicrobial activity, safety aspects, and some technological properties of bacteriocinogenic Enterococcus faecium from artisanal Tunisian fermented meat. Food Control 2010, 21, 462–470. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Anti-biofilm properties of the fecal probiotic Lactobacilli against Vibrio spp. Front. Cell. Infect. Microbiol. 2018, 8, 120. [Google Scholar] [CrossRef]
- Vidya, R.; Iyer, P. Antagonistic activity of probiotic organism against Vibrio cholerae and Cryptococcus neoformans. Malays. J. Microbiol. 2010, 6, 41–46. [Google Scholar]
- Simonetta, A.; Moragues de Velasco, L.; Frison, L. Antibacterial activity of enterococci strains against Vibrio cholerae. Lett. Appl. Microbiol. 1997, 24, 139–143. [Google Scholar] [CrossRef]
- Venkatesan, P.; Perfect, J.R.; Myers, S.A. Evaluation and management of fungal infections in immunocompromised patients. Dermatol. Ther. 2005, 18, 44–57. [Google Scholar] [CrossRef]
- Hedayati, M.; Pasqualotto, A.; Warn, P.; Bowyer, P.; Denning, D. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology 2007, 153, 1677–1692. [Google Scholar] [CrossRef]
- Bongomin, F.; Batac, C.; Richardson, M.D.; Denning, D.W. A review of onychomycosis due to Aspergillus species. Mycopathologia 2018, 183, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Baddley, J.W. Clinical risk factors for invasive aspergillosis. Med. Mycol. 2011, 49, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- IARC. Monographs on The Evaluation of Carcinogenic Risks to Humans; N°82. IARC: Some traditional herbal medicines, some mycotoxins, naphthalene and styrene; International Agency for Research on Cancer: Lyon, France, 2002. [Google Scholar]
- Kamei, K.; Watanabe, A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005, 43, 95–99. [Google Scholar] [CrossRef]
- Kuwaki, S.; Ohhira, I.; Takahata, M.; Murata, Y.; Tada, M. Antifungal activity of the fermentation product of herbs by lactic acid bacteria against tinea. J. Biosci. Bioeng. 2002, 94, 401–405. [Google Scholar] [CrossRef]
- Djaaboub, S.; Moussaoui, A.; Meddah, B.; Makhloufi, S.; Gouri, S.; El Khatib, R. Antifungal activity of some indigenous lactic acid bacteria isolated from soft wheat. J. Pure Appl. Microbiol. 2018, 12, 111–118. [Google Scholar] [CrossRef]
- Cortés-Zavaleta, O.; López-Malo, A.; Hernández-Mendoza, A.; García, H. Antifungal activity of lactobacilli and its relationship with 3-phenyllactic acid production. Int. J. Food Microbiol. 2014, 173, 30–35. [Google Scholar] [CrossRef]
- Sangmanee, P.; Hongpattarakere, T. Inhibitory of multiple antifungal components produced by Lactobacillus plantarum K35 on growth, aflatoxin production and ultrastructure alterations of Aspergillus flavus and Aspergillus parasiticus. Food Control 2014, 40, 224–233. [Google Scholar] [CrossRef]
- Belkacem-Hanfi, N.; Fhoula, I.; Semmar, N.; Guesmi, A.; Perraud-Gaime, I.; Ouzari, H.-I.; Boudabous, A.; Roussos, S. Lactic acid bacteria against post-harvest moulds and ochratoxin A isolated from stored wheat. Biol. Control 2014, 76, 52–59. [Google Scholar] [CrossRef]
- Marie, K.P.; Ngoufack, F.Z.; Edith, M.F.K.; Ciobotaru, O.; Matei, F.; Cornea, C.P.; Israel, R.F. Antifungal activity of lactic acid bacteria isolated from peanuts, gari, and orange fruit juice against food aflatoxigenic molds. Food Biotechnol. 2018, 32, 237–256. [Google Scholar] [CrossRef]
- Cosentino, S.; Viale, S.; Deplano, M.; Fadda, M.E.; Pisano, M.B. Application of autochthonous Lactobacillus strains as bio-preservatives to control fungal spoilage in caciotta cheese. BioMed Res. Int. 2018, 2018, 3915615. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, A.; Santiago, A.; Teixeira, J.A.; Venâncio, A.; Abrunhosa, L. Anti-aflatoxigenic effect of organic acids produced by Lactobacillus plantarum. Int. J. Food Microbiol. 2018, 264, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D. Antifungal activity of lactic acid bacteria isolated from Kimchi against Aspergillus fumigatus. Mycobiology 2005, 33, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Lasrado, L.D.; Gudipati, M. Antioxidant property of symbiotic combination of Lactobacillus sp. and wheat bran xylo-oligosaccharides. J. Food Sci. Technol. 2015, 52, 4551–4557. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, H.; Chen, W.; Zhong, Q.; Zhang, G.; Chen, W. Beneficial effects of tomato juice fermented by Lactobacillus plantarum and Lactobacillus casei: Antioxidation, antimicrobial effect, and volatile profiles. Molecules 2018, 23, 2366. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Proença, C.; Serralheiro, M.; Araujo, M. The in vitro screening for acetylcholinesterase inhibition and antioxidant activity of medicinal plants from Portugal. J. Ethnopharmacol. 2006, 108, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Yap, W.B.; Kofli, N.T.; Ghazali, A.R. Probiotic potentials of Lactobacillus plantarum isolated from fermented durian (Tempoyak), a Malaysian traditional condiment. Food Sci. Nutr. 2018, 6, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Abdhul, K.; Ganesh, M.; Shanmughapriya, S.; Kanagavel, M.; Anbarasu, K.; Natarajaseenivasan, K. Antioxidant activity of exopolysaccharide from probiotic strain Enterococcus faecium (BDU7) from Ngari. Int. J. Biol. Macromol. 2014, 70, 450–454. [Google Scholar] [CrossRef]
- Kaizu, H.; Sasaki, M.; Nakajima, H.; Suzuki, Y. Effect of antioxidative lactic acid bacteria on rats fed a diet deficient in vitamin E. J. Dairy Sci. 1993, 76, 2493–2499. [Google Scholar] [CrossRef]
- Parrella, A.; Caterino, E.; Cangiano, M.; Criscuolo, E.; Russo, C.; Lavorgna, M.; Isidori, M. Antioxidant properties of different milk fermented with lactic acid bacteria and yeast. Int. J. Food Sci. Technol. 2012, 47, 2493–2502. [Google Scholar] [CrossRef]
- Das, D.; Goyal, A. Antioxidant activity and γ-aminobutyric acid (GABA) producing ability of probiotic Lactobacillus plantarum DM5 isolated from Marcha of Sikkim. LWT Food Sci. Technol. 2015, 61, 263–268. [Google Scholar] [CrossRef]
- Prosekov, A.Y.; Dyshlyuk, L.S.; Milentieva, I.S.; Sykhikh, S.A.; Babich, O.O.; Ivanova, S.A.; Pavsky, V.A.; Shishin, M.V.; Matskova, L.V. Antioxidant and antimicrobial activity of bacteriocin-producing strains of lactic acid bacteria isolated from the human gastrointestinal tract. Prog. Nutr. 2017, 19, 67–80. [Google Scholar]
- Lin, M.Y.; Yen, C.L. Antioxidative ability of lactic acid bacteria. J. Agric. Food Chem. 1999, 47, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Okeke, B.C.; Andreazza, R.; Brandelli, A. Evaluation of selenite bioremoval from liquid culture by Enterococcus species. Microbiol. Res. 2011, 166, 176–185. [Google Scholar] [CrossRef]
- Divyashri, G.; Krishna, G.; Prapulla, S. Probiotic attributes, antioxidant, anti-inflammatory and neuromodulatory effects of Enterococcus faecium CFR 3003: In vitro and in vivo evidence. J. Med. Microbiol. 2015, 64, 1527–1540. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Okeke, B.C.; Camargo, F.A.d.O.; Brandelli, A. Antimicrobial and antioxidant activities of Enterococcus species isolated from meat and dairy products. Braz. J. Microbiol. 2015, 75, 923–931. [Google Scholar] [CrossRef]
| LAB Strain | n-Hexadecane | Chloroform | Ethyl Acetate |
|---|---|---|---|
| L. paraplantarum A1 | 90 ± 0.4 a (H§) | 53 ± 0.2 bc (M§) | 64 ± 0.2 a (M) |
| L. plantarum A2 | 90 ± 0.4 a (H) | 70 ± 0.5 a (H) | 66± 0.0 a (M) |
| L. plantarum A3 | 79 ± 0.5 b (H) | 70 ± 1.2 a (H) | 24 ± 2.1 c (M) |
| E. faecalis A4 | 64 ± 1.2 b (M) | 66 ± 1.2 ab (M) | 39 ± 0.6 bc (M) |
| E. faecalis A5 | 55 ± 1.3 c (M) | 73 ± 0.6 a (H) | 51 ± 1.2 a (M) |
| E. faecalis A6 | 75 ± 0.6 b (H) | 65 ± 0.9 ab (M) | 35 ± 0.3 bc (L§) |
| E. faecalis A7 | 83 ± 0.5 ab (H) | 71 ± 0.8 a (H) | 21 ± 2.7 c (L) |
| E. faecalis A8 | 78 ± 1.2 b (H) | 61 ± 0.3 b (M) | 24 ± 0.6 c (L) |
| E. faecalis A9 | 75 ± 0.9 b (H) | 73 ± 0.7 a (H) | 8 ± 0.8 d (L) |
| E. faecalis A10 | 85 ± 0.3 ab (H) | 46 ± 0.2 c (M) | 22 ± 0.4 c (L) |
| E. faecalis A11 | 77 ± 0.5 b (H) | 59 ± 1.0 bc (M) | 46 ± 0.4 b (M) |
| E. faecalis A12 | 90 ± 0.4 a (H) | 53 ± 0.2 bc (M) | 64 ± 0.2 a (M) |
| E. faecalis A13 | 90 ± 0.4 a (H) | 71 ± 0.5 a (H) | 66 ± 0.0 a (M) |
| E. faecalis A14 | 79 ± 0.5 b (H) | 70 ± 1.2 a (H) | 24 ± 2.1 c (M) |
| LAB Strain | Aspergillus flavus | Aspergillus carbonarius | I% of A. flavus | I% of A. carbonarius | ||
|---|---|---|---|---|---|---|
| GI% | GI Category | GI% | GI Category | |||
| A1 | 50 ± 3.5 b | 2 | 97 ± 0.6 a | 4 | 40 ± 1.5 b | 67 ± 2.0 b |
| A2 | 52 ± 3.4 b | 3 | 85 ± 0.8 a | 4 | 28 ± 1.7 b | 80 ± 1.0 b |
| A3 | 50 ± 3.5 b | 3 | 90 ± 0.6 a | 4 | 25 ± 2.0 b | 78 ±1.3 b |
| A4 | 56 ± 2.6 b | 3 | 89 ± 1.4 a | 4 | 61 ± 1.1 a | 94 ± 2.9 a |
| A5 | 63 ± 2.1 ab | 3 | 89 ± 1.3 a | 4 | 59 ± 1.4 a | 95 ± 1.1 a |
| A6 | 56 ± 2.5 b | 3 | 89 ± 1.3 a | 4 | 58 ± 1.0 a | 95 ± 1.1 a |
| A7 | 62 ± 1.9 ab | 3 | 87 ± 1.0 a | 4 | 58 ± 9.4 a | 95 ± 1.4 a |
| A8 | 68 ± 1.7 a | 3 | 77 ± 2.0 abc | 4 | 57 ± 1.1 a | 95 ± 1.6 a |
| A9 | 66 ± 2.0 a | 3 | 81 ± 1.6 a | 4 | 62 ± 1.2 a | 94 ± 1.3 a |
| A10 | 70 ± 1.7 a | 3 | 86 ± 1.4 a | 4 | 58 ± 1.1 a | 95 ± 1.1 a |
| A11 | 70 ± 1.7 a | 3 | 87 ± 1.3 a | 4 | 56 ± 1.2 a | 95 ± 1.1 a |
| A12 | 53 ± 2.9 b | 3 | 71 ± 2.3 bc | 3 | 54 ± 1.1 a | 95 ±0,9 a |
| A13 | 71 ± 1.6 a | 3 | 83 ± 1.4 a | 4 | 51 ± 9.7 a | 94 ± 1.2 a |
| A14 | 68 ± 1.8 a | 3 | 59 ± 2.5 c | 3 | 54 ± 1.1 a | 96 ± 1.0 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Missaoui, J.; Saidane, D.; Mzoughi, R.; Minervini, F. Fermented Seeds (“Zgougou”) from Aleppo Pine as a Novel Source of Potentially Probiotic Lactic Acid Bacteria. Microorganisms 2019, 7, 709. https://doi.org/10.3390/microorganisms7120709
Missaoui J, Saidane D, Mzoughi R, Minervini F. Fermented Seeds (“Zgougou”) from Aleppo Pine as a Novel Source of Potentially Probiotic Lactic Acid Bacteria. Microorganisms. 2019; 7(12):709. https://doi.org/10.3390/microorganisms7120709
Chicago/Turabian StyleMissaoui, Jihen, Dalila Saidane, Ridha Mzoughi, and Fabio Minervini. 2019. "Fermented Seeds (“Zgougou”) from Aleppo Pine as a Novel Source of Potentially Probiotic Lactic Acid Bacteria" Microorganisms 7, no. 12: 709. https://doi.org/10.3390/microorganisms7120709
APA StyleMissaoui, J., Saidane, D., Mzoughi, R., & Minervini, F. (2019). Fermented Seeds (“Zgougou”) from Aleppo Pine as a Novel Source of Potentially Probiotic Lactic Acid Bacteria. Microorganisms, 7(12), 709. https://doi.org/10.3390/microorganisms7120709





