Characterization and Phylodynamics of Reassortant H12Nx Viruses in Northern Eurasia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, Virus Isolation, and Cells
2.2. Experimental Infection of Chickens and Mice
2.3. Antigenic Analysis
2.4. Susceptibility to Neuraminidase Inhibitors
2.5. Sequencing
2.6. Genetic Analysis
3. Results
3.1. Sampling and Virus Isolation
3.2. Virological Characteristics
3.3. Antigenic Analysis
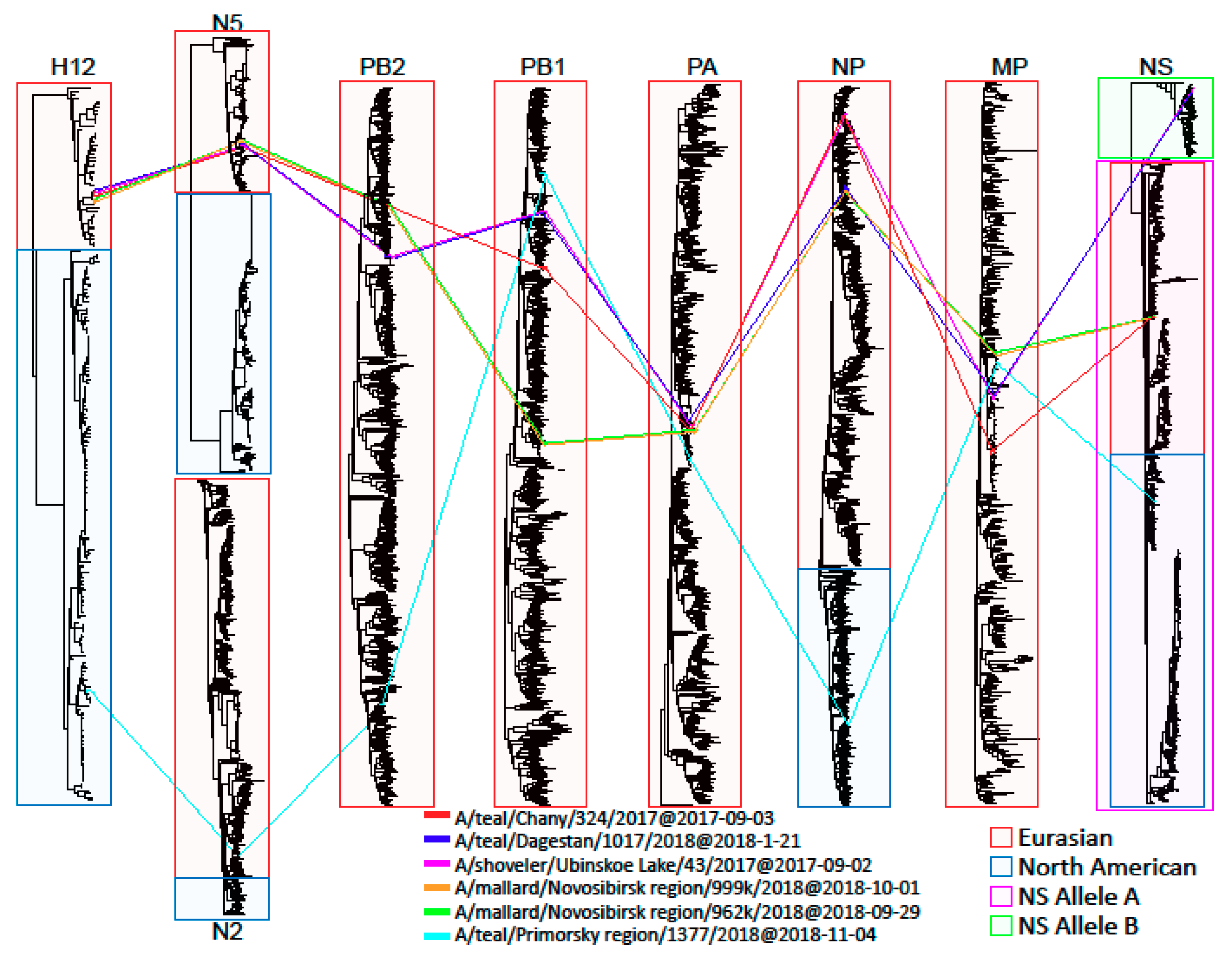
3.4. Genetic Analysis
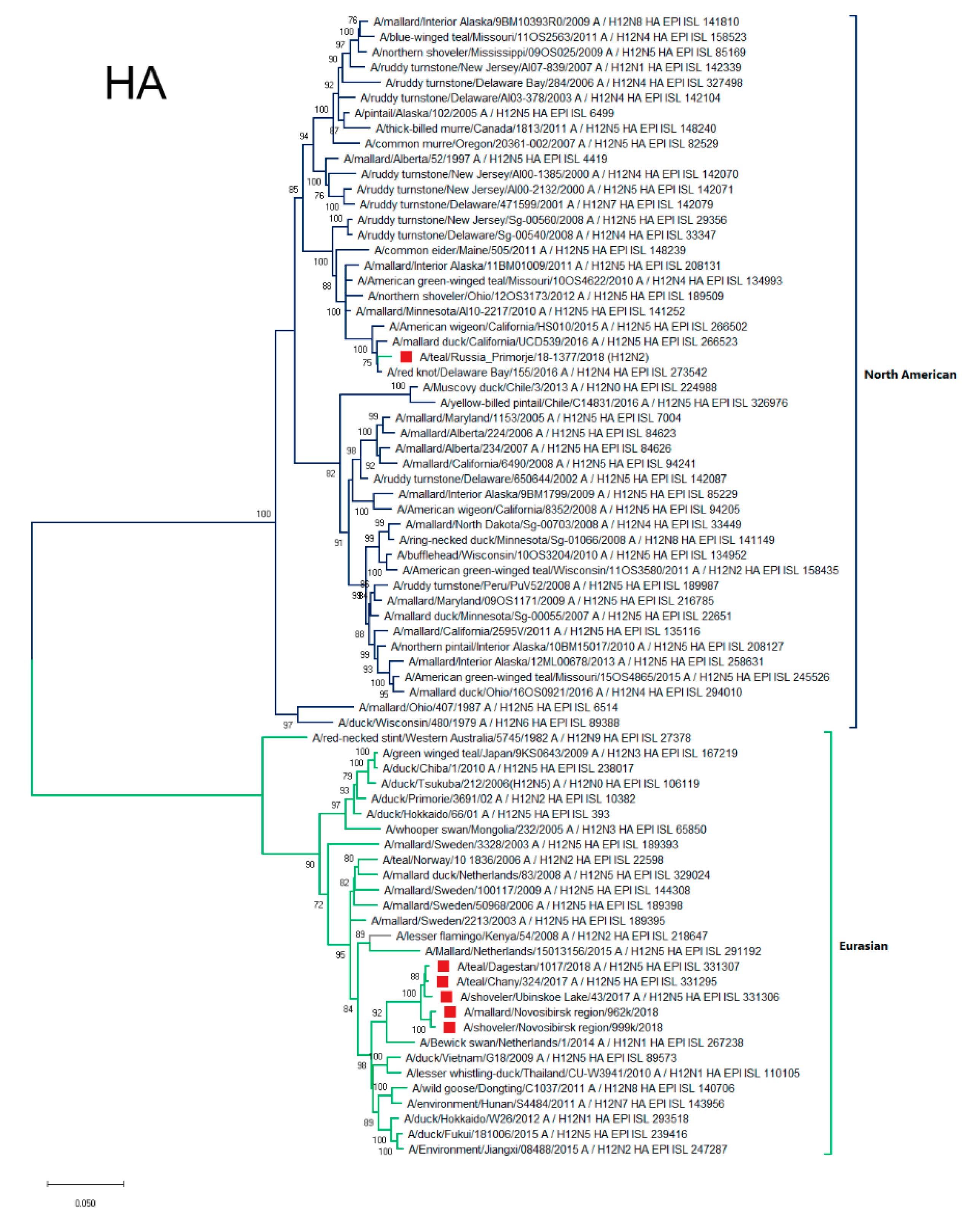
3.4.1. HA
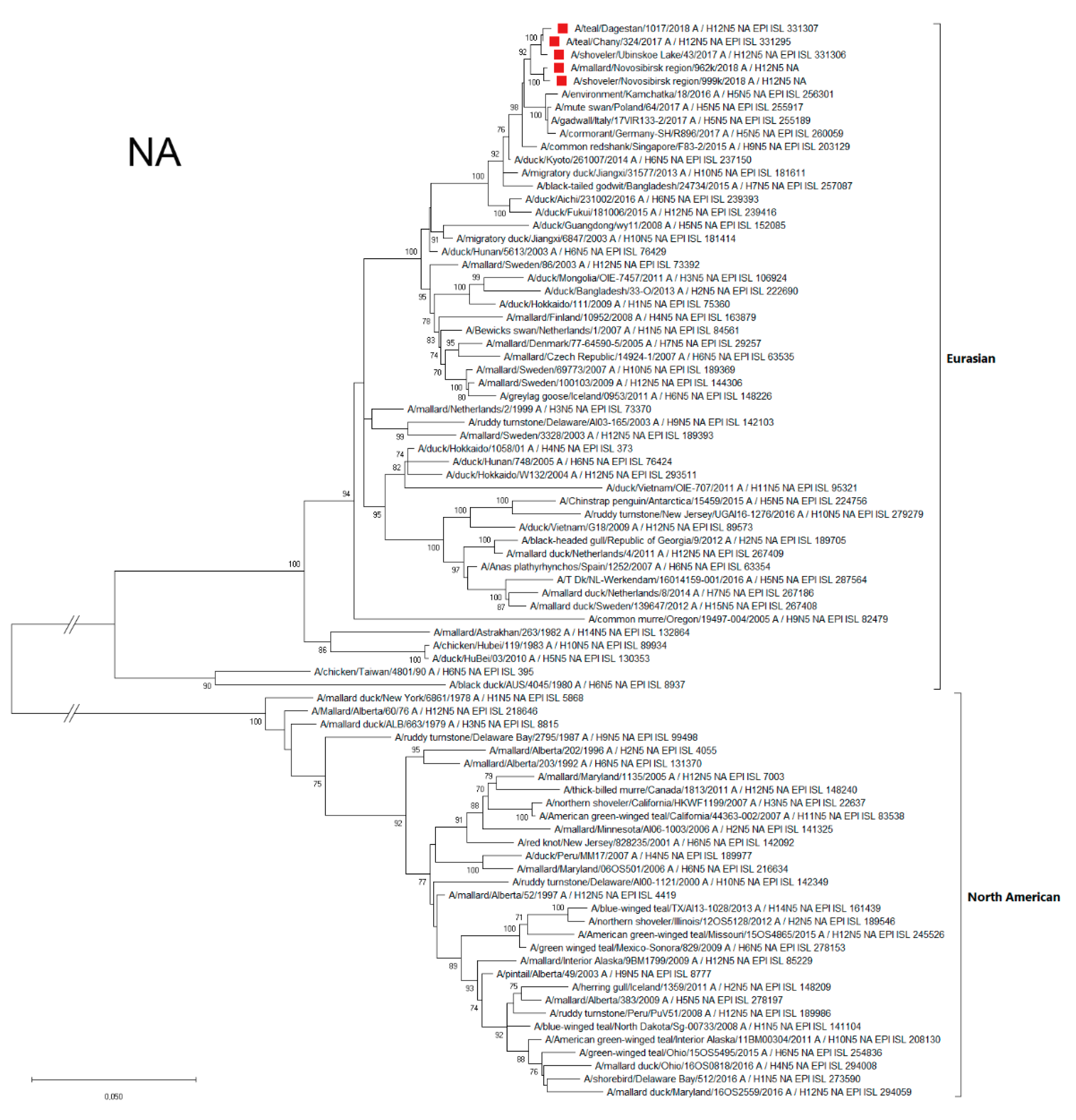
3.4.2. NA
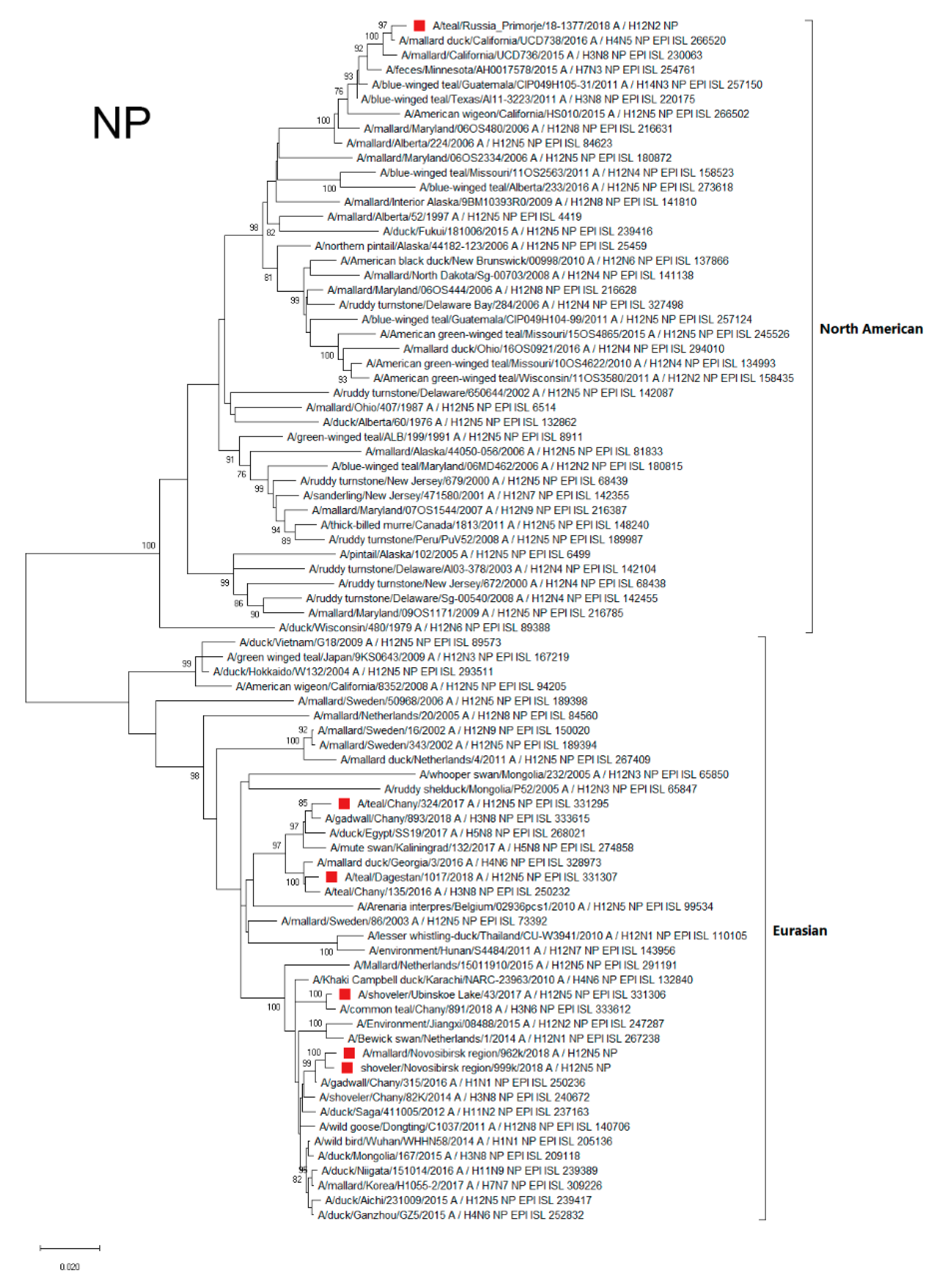
3.4.3. NP
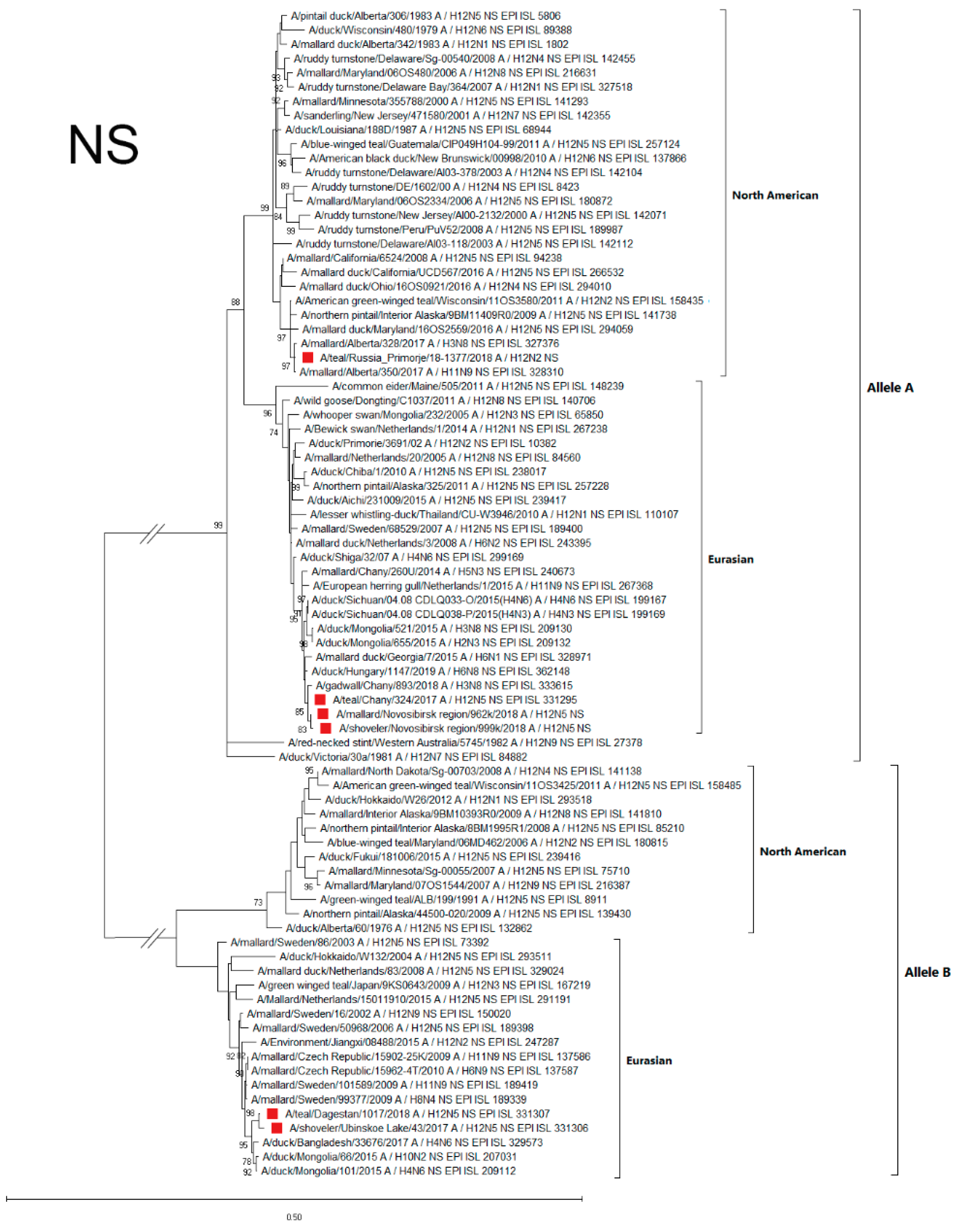
3.4.4. NS
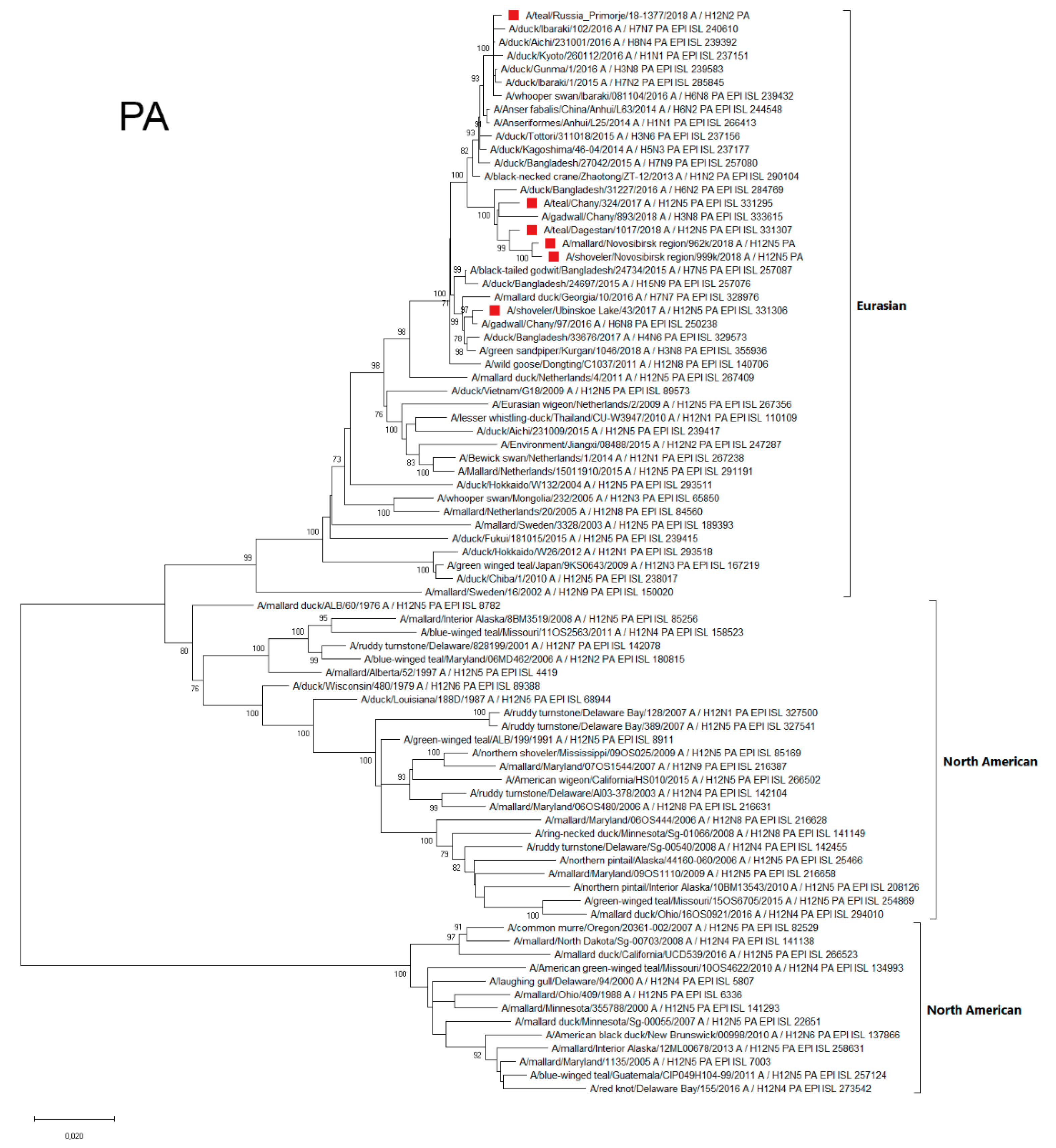
3.4.5. PA
3.4.6. PB1
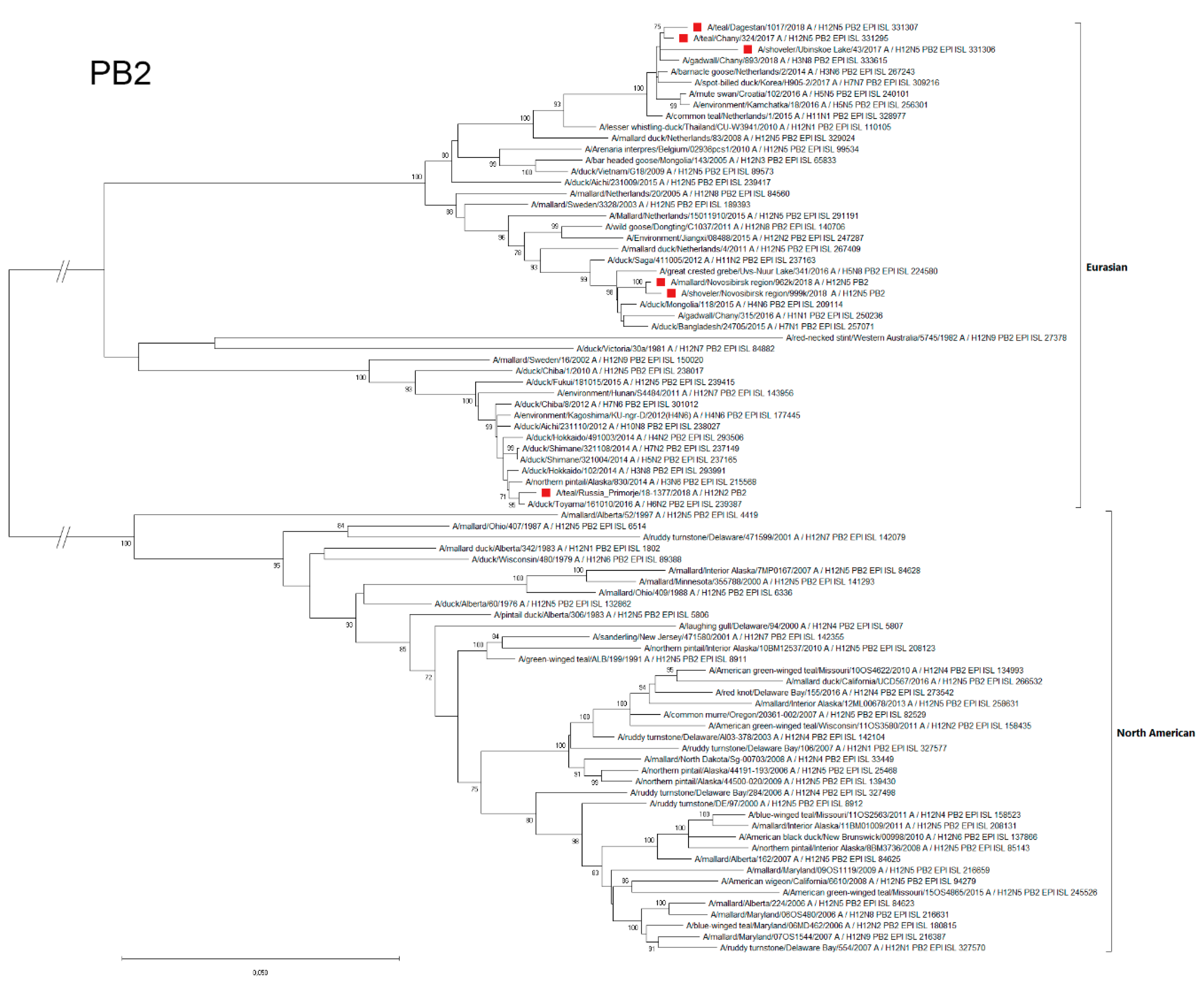
3.4.7. PB2
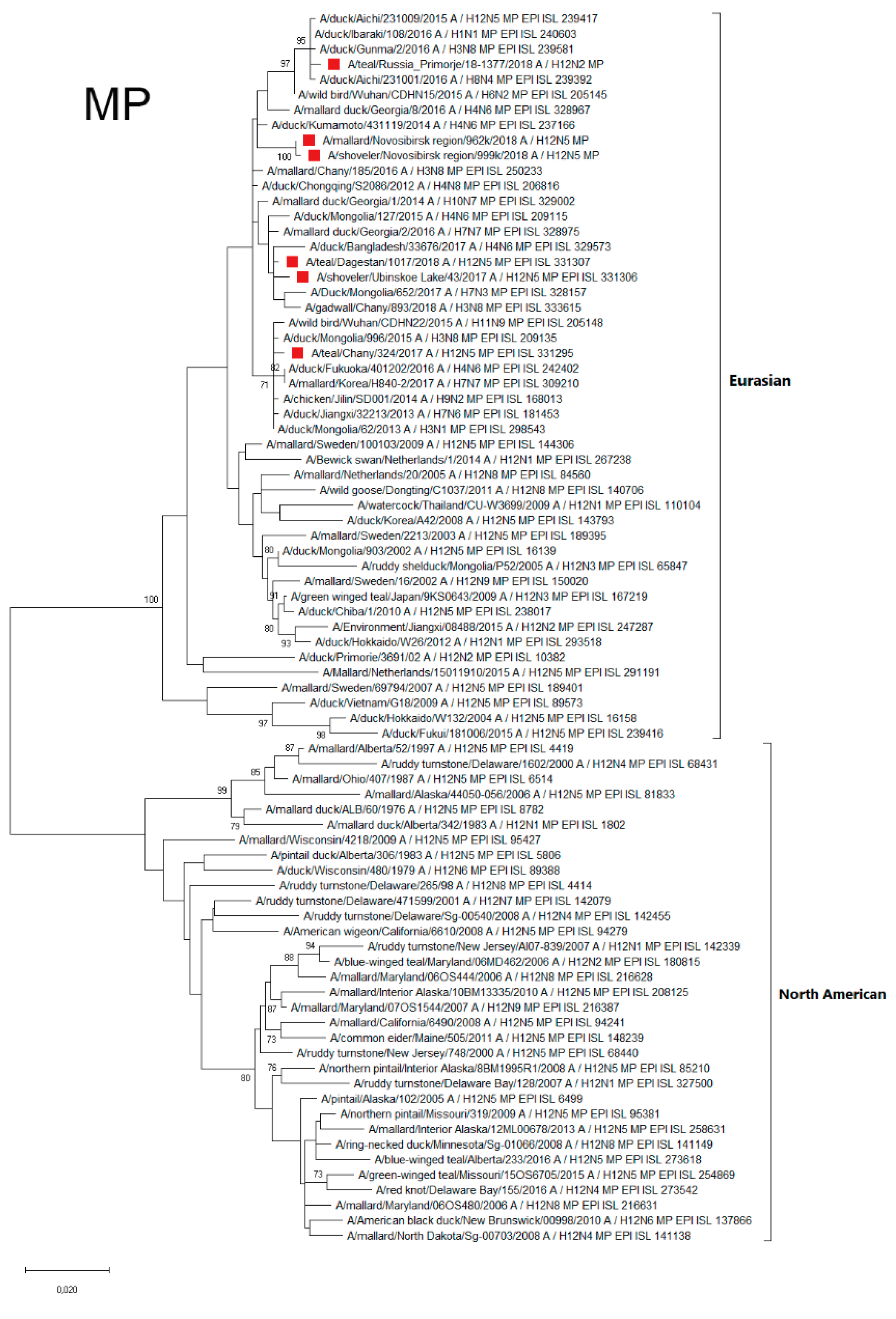
3.4.8. MP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.A.; Chen, L.-M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza a virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza A viruses. Curr. Top Microbiol. Immunol. 2014, 385, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Antigua, K.J.C.; Choi, W.S.; Baek, Y.H.; Song, M.S. The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx. Microorganisms 2019, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Veits, J.; Mettenleiter, T.C. Prevalence and control of H7 avian influenza viruses in birds and humans. Epidemiol. Infect. 2014, 142, 896–920. [Google Scholar] [CrossRef]
- Veits, J.; Weber, S.; Stech, O.; Breithaupt, A.; Gräber, M.; Gohrbandt, S.; Bogs, J.; Hundt, J.; Teifke, J.P.; Mettenleiter, T.C.; et al. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc. Natl. Acad. Sci. USA 2012, 109, 2579–2584. [Google Scholar] [CrossRef]
- Wille, M.; Latorre-Margalef, N.; Tolf, C.; Halpin, R.; Wentworth, D.; Fouchier, R.A.M.; Raghwani, J.; Pybus, O.G.; Olsen, B.; Waldenström, J. Where do all the subtypes go? Temporal dynamics of H8-H12 influenza a viruses in waterfowl. Virus Evol. 2018, 4, vey025. [Google Scholar] [CrossRef]
- Sharp, G.B.; Kawaoka, Y.; Wright, S.M.; Turner, B.; Hinshaw, V.; Webster, R.G. Wild ducks are the reservoir for only a limited number of influenza a subtypes. Epidemiol. Infect. 1993, 110, 161. [Google Scholar] [CrossRef]
- De Marco, M.A.; Sharshov, K.; Gulyaeva, M.; Delogu, M.; Ciccarese, L.; Castrucci, M.R.; Shestopalov, A. Ecology of Avian Influenza Viruses in Siberia. In Siberia: Ecology, Diversity and Environmental Impact; Nova Science Pub Inc.: Hauppauge, NY, USA, 2016; pp. 83–160. [Google Scholar]
- Scott, D.A.; Rose, P.M. Wetlands International Atlas of the Anatidae Populations in Africa and Western Eurasia; Wetlands International: Wageningen, The Netherlands, 1996; ISBN 978-1-900442-09-1. [Google Scholar]
- Horman, W.S.J.; Nguyen, T.H.O.; Kedzierska, K.; Bean, A.G.D.; Layton, D.S. The Drivers of Pathology in Zoonotic Avian Influenza: The Interplay between Host and Pathogen. Front. Immunol. 2018, 9, 1812. [Google Scholar] [CrossRef]
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic Potential of Influenza a Viruses: A Comprehensive Overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef]
- Spackman, E.; Killian, M.L. Avian Influenza Virus Isolation, Propagation, and Titration in Embryonated Chicken Eggs. In Animal Influenza Virus; Methods in Molecular Biology; Spackman, E., Ed.; Springer: New York, NY, USA, 2014; pp. 125–140. ISBN 978-1-4939-0758-8. [Google Scholar]
- Terrestrial Code: OIE -World Organisation for Animal Health. Available online: https://www.oie.int/international-standard-setting/terrestrial-code/ (accessed on 5 September 2019).
- World Health Organization. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization: Geneva, Switzerland, 2011; ISBN 978-92-4-154809-0. [Google Scholar]
- Mine, J.; Uchida, Y.; Sharshov, K.; Sobolev, I.; Shestopalov, A.; Saito, T. Phylogeographic evidence for the inter- and intracontinental dissemination of avian influenza viruses via migration flyways. PLoS ONE 2019, 14, e0218506. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Welch, J.J.; Suchard, M.A. Phylogeography Takes a Relaxed Random Walk in Continuous Space and Time. Mol. Biol. Evol. 2010, 27, 1877–1885. [Google Scholar] [CrossRef]
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol. Biol. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef]
- Andraszewicz, S.; Scheibehenne, B.; Rieskamp, J.; Grasman, R.; Verhagen, J.; Wagenmakers, E.J. An Introduction to Bayesian Hypothesis Testing for Management Research. J. Manag. 2015, 41, 521–543. [Google Scholar]
- Zohari, S.; Gyarmati, P.; Ejdersund, A.; Berglöf, U.; Thorén, P.; Ehrenberg, M.; Czifra, G.; Belák, S.; Waldenström, J.; Olsen, B.; et al. Phylogenetic analysis of the non-structural (NS) gene of influenza a viruses isolated from mallards in Northern Europe in 2005. Virol. J. 2008, 5, 147. [Google Scholar] [CrossRef]
- Schrauwen, E.J.A.; De Graaf, M.; Herfst, S.; Rimmelzwaan, G.F.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Determinants of virulence of influenza a virus. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 479–490. [Google Scholar] [CrossRef]
- Nobusawa, E.; Aoyama, T.; Kato, H.; Suzuki, Y.; Tateno, Y.; Nakajima, K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza a viruses. Virology 1991, 182, 475–485. [Google Scholar] [CrossRef]
- Ha, Y.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. USA 2001, 98, 11181–11186. [Google Scholar] [CrossRef]
- Bui, V.N.; Ogawa, H.; Hussein, I.T.M.; Hill, N.J.; Trinh, D.Q.; AboElkhair, M.; Sultan, S.; Ma, E.; Saito, K.; Watanabe, Y.; et al. Genetic characterization of a rare H12N3 avian influenza virus isolated from a green-winged teal in Japan. Virus Genes 2015, 50, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Leung, B.W.; Chen, H.; Brownlee, G.G. Correlation between polymerase activity and pathogenicity in two duck H5N1 influenza viruses suggests that the polymerase contributes to pathogenicity. Virology 2010, 401, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Hule-Post, D.J.; Franks, J.; Boyd, K.; Salomon, R.; Hoffmann, E.; Yen, H.L.; Webby, R.J.; Walker, D.; Nguyen, T.D.; Webster, R.G. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 2007, 81, 8515–8524. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hu, W.B.; Xu, K.; He, Y.X.; Wang, T.Y.; Chen, Z.; Li, T.X.; Liu, J.H.; Buchy, P.; Sun, B. Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J. Gen. Virol. 2012, 93, 531–540. [Google Scholar] [CrossRef]
- Schmolke, M.; Manicassamy, B.; Pena, L.; Sutton, T.; Hai, R.; Varga, Z.T.; Hale, B.G.; Steel, J.; Pérez, D.R.; García-Sastre, A. Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. PLoS Pathog. 2011, 7, e1002186. [Google Scholar] [CrossRef]
- Salomon, R.; Franks, J.; Govorkova, E.A.; Ilyushina, N.A.; Yen, H.L.; Hulse-Post, D.J.; Humberd, J.; Trichet, M.; Rehg, J.E.; Webby, R.J.; et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 2006, 203, 689–697. [Google Scholar] [CrossRef]
- Bogs, J.; Kalthoff, D.; Veits, J.; Pavlova, S.; Schwemmle, M.; Mänz, B.; Mettenleiter, T.C.; Stech, J. Reversion of PB2-627E to -627K during replication of an H5N1 Clade 2.2 virus in mammalian hosts depends on the origin of the nucleoprotein. J. Virol. 2011, 85, 10691–10698. [Google Scholar] [CrossRef]
- Li, Z.; Chen, H.; Jiao, P.; Deng, G.; Tian, G.; Li, Y.; Hoffmann, E.; Webster, R.G.; Matsuoka, Y.; Yu, K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005, 79, 12058–12064. [Google Scholar] [CrossRef]
- Li, J.; Ishaq, M.; Prudence, M.; Xi, X.; Hu, T.; Liu, Q.; Guo, D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009, 144, 123–129. [Google Scholar] [CrossRef]
- Kawaoka, Y.; Yamnikova, S.; Chambers, T.M.; Lvov, D.K.; Webster, R.G. Molecular characterization of a new hemagglutinin, subtype H14, of influenza a virus. Virology 1990, 179, 759–767. [Google Scholar] [CrossRef]
- Sivay, M.V.; Sayfutdinova, S.G.; Sharshov, K.A.; Alekseev, A.Y.; Yurlov, A.K.; Runstadler, J.; Shestopalov, A.M. Surveillance of Influenza a Virus in Wild Birds in the Asian Portion of Russia in 2008. Avdi 2012, 56, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Muzyka, D.; Pantin-Jackwood, M.; Spackman, E.; Smith, D.; Rula, O.; Muzyka, N.; Stegniy, B. Isolation and Genetic Characterization of Avian Influenza Viruses Isolated from Wild Birds in the Azov-Black Sea Region of Ukraine (2001–2012). Avian Dis. 2016, 60, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152. [Google Scholar] [PubMed]
- Wille, M.; Robertson, G.J.; Whitney, H.; Bishop, M.A.; Runstadler, J.A.; Lang, A.S. Extensive Geographic Mosaicism in Avian Influenza Viruses from Gulls in the Northern Hemisphere. PLoS ONE 2011, 6, e20064. [Google Scholar] [CrossRef]
- Nagy, A.; Mettenleiter, T.C.; Abdelwhab, E.M. A brief summary of the epidemiology and genetic relatedness of avian influenza H9N2 virus in birds and mammals in the Middle East and North Africa. Epidemiol. Infect. 2017, 145, 3320–3333. [Google Scholar] [CrossRef]
- Chen, R.; Holmes, E.C. Frequent inter-species transmission and geographic subdivision in avian influenza viruses from wild birds. Virology 2009, 383, 156–161. [Google Scholar] [CrossRef][Green Version]
- Isakov, Y.A. MAR Project and Conservation of Waterfowl Breeding in the USSR; Salverda, Z., Ed.; IWRB: Noordwijk aan zee, The Netherlands, 1966; pp. 125–138. [Google Scholar]
- Veen, J.; Yurlov, A.K.; Delany, S.N.; Mihantiev, A.I.; Selivanova, M.A.; Boere, G.C. An Atlas of Movements of Southwest Siberian Waterbirds; Wetlands International: Wageningen, The Netherlands, 2005. [Google Scholar]
- Boere, G.G. Waterbirds around the World: A Global Overview of the Conservation, Management and Research of the World’s Waterbird Flyways; Scottish Natural Heritage: Edinburgh, Scotland, 2007; ISBN 978-0-11-497333-9. [Google Scholar]

| Viruses | Sampling Date | Sampling Region/ Sample Size at Site | Host Species (Latin Name) | Host Species (English Name) | Virus Subtype | Strain Name |
|---|---|---|---|---|---|---|
| A/43 | September 17 | Western Siberia/590 | Anas clypeata | Shoveler | H12N5 | A/shoveler/Ubinskoe Lake/43/2017 |
| A/324 | September 17 | Anas crecca | Common teal | H12N5 | A/teal/Chany/324/2017 | |
| A/1017 | January18 | Caspian region/304 | Anas crecca | Common teal | H12N5 | A/teal/Dagestan/1017/2018 |
| A/962 | September 18 | Western Siberia/478 | Anas platyrhynchos | Mallard | H12N5 | A/mallard/Novosibirsk region/962k/2018 |
| A/999 | September 18 | Anas clypeata | Shoveler | H12N5 | A/shoveler/Novosibirsk region/999k/2018 | |
| A/1377-Amer | November 18 | Far East/280 | Anas crecca | Common teal | H12N2 | A/teal/Russia_Primorje/18-1377/2018 |
| Viruses | log10TCID50/mL | log10EID50/mL | IVPI | Pathogenicity for Mice | Oseltamivir Carboxylate IC50 (nM) | Phenotype b |
|---|---|---|---|---|---|---|
| A/43 | 5.4 ± 0.3 | 8.3 ± 0.3 | 0 | np a | 12.47 | RI |
| A/324 | 5.6 ± 0.3 | 8.3 ± 0.2 | 0 | np | 7.5 | S |
| A/1017 | 5.0 ± 0.2 | 7.9 ± 0.4 | 0 | np | 9.2 | S |
| A/962 | 5.4 ± 0.2 | 8.0 ± 0.2 | 0 | np | 0.4 | S |
| A/999 | 5.8 ± 0.3 | 7.8 ± 0.4 | 0 | np | 0.4 | S |
| A/1377 | 5.3 ± 0.2 | 8.0 ± 0.3 | 0 | np | 4.4 | S |
| Chicken Post Infectious Sera | Antigens | |||||
|---|---|---|---|---|---|---|
| A/43 | A/324 | A/1017 | A/962 | A/999 | A/1377 | |
| A/43 | 160 | 160 | 80 | 160 | 320 | 80 |
| A/324 | 160 | 160 | 80 | 160 | 640 | 160 |
| A/1017 | 160 | 160 | 160 | 320 | 640 | 160 |
| A/962 | 80 | 80 | 80 | 160 | 160 | 40 |
| A/999 | 80 | 80 | 80 | 80 | 160 | 40 |
| A/1377-Amer | 80 | 80 | 80 | 80 | 160 | 160 |
| Gene | Amino Acid Site | Strain | Effect | Subtype Showed to Be Affected | Reference |
|---|---|---|---|---|---|
| PA | 149S | A/1377, A/324, A/962, A/999, A/1017, A/43 | P149S—limited lethality in mice | H5N1 | [28] |
| PA | 515A | A/1377, A/324, A/962, A/999, A/1017, A/43 | T515A—polymerase activity decreasing | H5N1 | [29] |
| PB1 | 598L | A/1377, A/324, A/962, A/999, A/1017, A/43 | P598L—replication decreasing in MDCK | H1N1, H5N1 | [30] |
| PB1-F2 | 66S | A/962, A/999 | N66S—replication increasing | H5N1 | [31] |
| PB2 | 553V | A/324 | I553V—polymerase activity decreasing | H5N1 | [28] |
| PB2 | 391E, 627E | A/1377, A/324, A/962, A/999, A/1017, A/43 | Q391E—virulence decreasing in ferrets; K627E—replication decreasing in mammalian cells | H5N1 | [32,33] |
| PB2 | 701D | A/1377, A/324, A/962, A/999, A/1017 | N701D—lethality increasing in mice | H5N1 | [34] |
| PB2 | 89V, 309D, 339K, 477G, 495V, 627E, 676T 1 | A/1377, A/324, A/962, A/999, A/1017, A/43 | L89V, G309D, T339K, R477G, I495V, K627E, A676T—polymerase activity increasing in mouse cells | H5N1 | [35] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharshov, K.; Mine, J.; Sobolev, I.; Kurskaya, O.; Dubovitskiy, N.; Kabilov, M.; Alikina, T.; Nakayama, M.; Tsunekuni, R.; Derko, A.; et al. Characterization and Phylodynamics of Reassortant H12Nx Viruses in Northern Eurasia. Microorganisms 2019, 7, 643. https://doi.org/10.3390/microorganisms7120643
Sharshov K, Mine J, Sobolev I, Kurskaya O, Dubovitskiy N, Kabilov M, Alikina T, Nakayama M, Tsunekuni R, Derko A, et al. Characterization and Phylodynamics of Reassortant H12Nx Viruses in Northern Eurasia. Microorganisms. 2019; 7(12):643. https://doi.org/10.3390/microorganisms7120643
Chicago/Turabian StyleSharshov, Kirill, Junki Mine, Ivan Sobolev, Olga Kurskaya, Nikita Dubovitskiy, Marsel Kabilov, Tatiana Alikina, Momoko Nakayama, Ryota Tsunekuni, Anastasiya Derko, and et al. 2019. "Characterization and Phylodynamics of Reassortant H12Nx Viruses in Northern Eurasia" Microorganisms 7, no. 12: 643. https://doi.org/10.3390/microorganisms7120643
APA StyleSharshov, K., Mine, J., Sobolev, I., Kurskaya, O., Dubovitskiy, N., Kabilov, M., Alikina, T., Nakayama, M., Tsunekuni, R., Derko, A., Prokopyeva, E., Alekseev, A., Shchelkanov, M., Druzyaka, A., Gadzhiev, A., Uchida, Y., Shestopalov, A., & Saito, T. (2019). Characterization and Phylodynamics of Reassortant H12Nx Viruses in Northern Eurasia. Microorganisms, 7(12), 643. https://doi.org/10.3390/microorganisms7120643







