Linking Soil Fungal Generality to Tree Richness in Young Subtropical Chinese Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site
2.2. Soil Sampling
2.3. Nucleic Acid Extraction and Multiplexed Amplicon Pyrosequencing
2.4. Bioinformatic Analysis
2.5. Data Processing
2.6. Tree-Fungal Bipartite Analysis in a Subsampling Approach
2.7. Specialization Analysis
3. Results
3.1. Tree-Fungal Bipartite Network Analysis with a Subsampling Approach
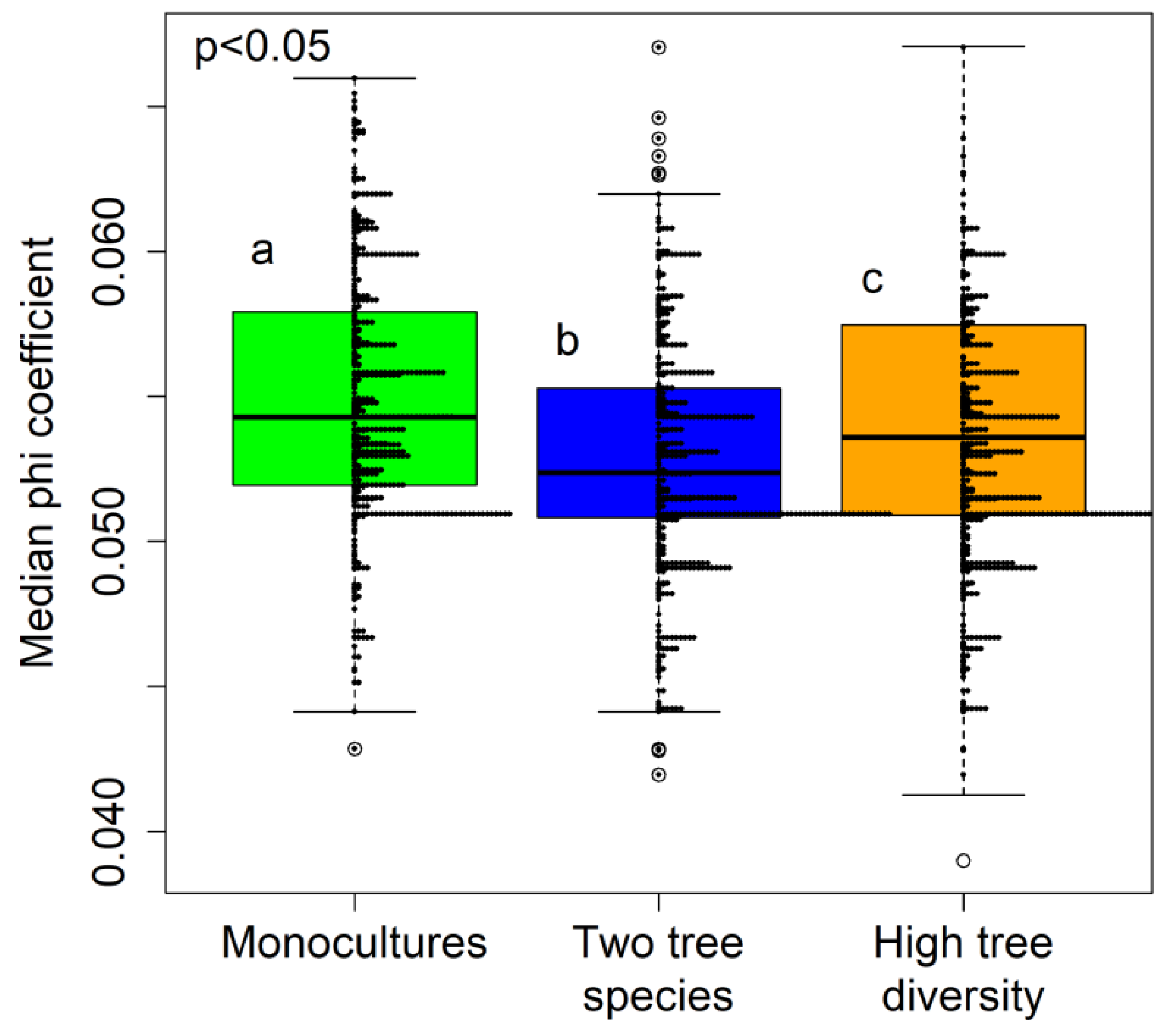
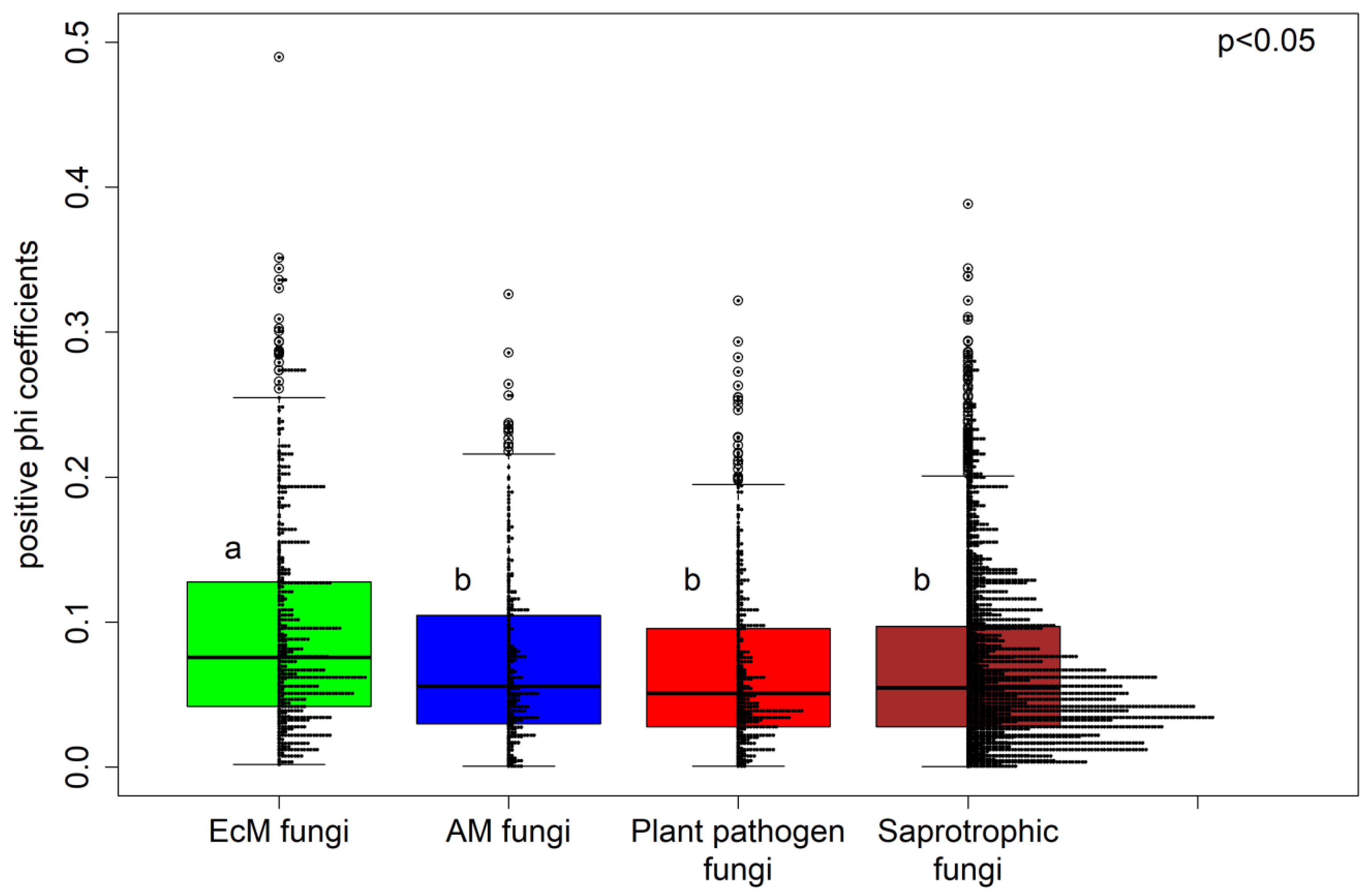
3.2. Fungal Specialization Patterns as Evaluated using the Phi Coefficient
4. Discussion
4.1. Increased Fungal Alpha Diversity in Plots with High Tree Species Diversity
4.2. The Connectance and Fungal Generality of Tree-Fungal Bipartite Networks are Highest at the Two Tree Species Diversity Level
4.3. Comparison with Other Bipartite Network Studies
5. Conclusions
Data availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blackwell, M. The fungi: 1, 2, 3… 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial diversity in soil: Selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Herold, N.; Schöning, I.; Gutknecht, J.; Alt, F.; Boch, S.; Müller, J.; Oelmann, Y.; Socher, S.A.; Wilcke, W.; Wubet, T.; et al. Soil property and management effects on grassland microbial communities across a latitudinal gradient in germany. Appl. Soil Ecol. 2014, 73, 41–50. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Schappe, T.; Albornoz, F.E.; Turner, B.L.; Neat, A.; Condit, R.; Jones, F.A. The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three panamanian rainforests. J. Ecol. 2017, 105, 569–579. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal Ruiz, L.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Fungal biogeography. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Saitta, A.; Anslan, S.; Bahram, M.; Brocca, L.; Tedersoo, L. Tree species identity and diversity drive fungal richness and community composition along an elevational gradient in a mediterranean ecosystem. Mycorrhiza 2018, 28, 39–47. [Google Scholar] [CrossRef]
- Urbanova, M.; Snajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Weißbecker, C.; Wubet, T.; Lentendu, G.; Kühn, P.; Scholten, T.; Bruelheide, H.; Buscot, F. Experimental evidence of functional group-dependent effects of tree diversity on soil fungi in subtropical forests. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.E.; Raich, J.W.; Valverde-Barrantes, O.J.; Fisher, R.F. Tree species effects on soil properties in experimental plantations in tropical moist forest. Soil Sci. Soc. Am. J. 2007, 71, 1389–1397. [Google Scholar] [CrossRef]
- Menyailo, O.V.; Hungate, B.A.; Zech, W. Tree species mediated soil chemical changes in a siberian artificial afforestation experiment: Tree species and soil chemistry. Plant Soil 2002, 242, 171–182. [Google Scholar] [CrossRef]
- Reich, P.B.; Oleksyn, J.; Modrzynski, J.; Mrozinski, P.; Hobbie, S.E.; Eissenstat, D.M.; Chorover, J.; Chadwick, O.A.; Hale, C.M.; Tjoelker, M.G. Linking litter calcium, earthworms and soil properties: A common garden test with 14 tree species. Ecol. Lett. 2005, 8, 811–818. [Google Scholar] [CrossRef]
- Augusto, L.; Ranger, J.; Binkley, D.; Rothe, A. Impact of several common tree species of european temperate forests on soil fertility. Ann. For. Sci. 2002, 59, 233–253. [Google Scholar] [CrossRef]
- Aussenac, G. Interactions between forest stands and microclimate: Ecophysiological aspects and consequences for silviculture. Ann. For. Sci. 2000, 57, 287–301. [Google Scholar] [CrossRef]
- Sapijanskas, J.; Paquette, A.; Potvin, C.; Kunert, N.; Loreau, M. Tropical tree diversity enhances light capture through crown plasticity and spatial and temporal niche differences. Ecology 2014, 95, 2479–2492. [Google Scholar] [CrossRef]
- Williams, L.J.; Paquette, A.; Cavender-Bares, J.; Messier, C.; Reich, P.B. Spatial complementarity in tree crowns explains overyielding in species mixtures. Nat. Ecol. Evol. 2017, 1. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.J.; Schmid, B.; Bruelheide, H.; Bu, W.S.; Ma, K.P. Positive effects of tree species richness on fine-root production in a subtropical forest in se-china. J. Plant Ecol. 2017, 10, 146–157. [Google Scholar] [CrossRef]
- Brassard, B.W.; Chen, H.Y.H.; Cavard, X.; Laganiere, J.; Reich, P.B.; Bergeron, Y.; Pare, D.; Yuan, Z.Y. Tree species diversity increases fine root productivity through increased soil volume filling. J. Ecol. 2013, 101, 210–219. [Google Scholar] [CrossRef]
- Dimitrakopoulos, P.G.; Schmid, B. Biodiversity effects increase linearly with biotope space. Ecol. Lett. 2004, 7, 574–583. [Google Scholar] [CrossRef]
- Peers, M.J.L.; Thornton, D.H.; Murray, D.L. Reconsidering the specialist-generalist paradigm in niche breadth dynamics: Resource gradient selection by canada lynx and bobcat. PLoS ONE 2012, 7, e51488. [Google Scholar] [CrossRef] [PubMed]
- Burkle, L.A.; Marlin, J.C.; Knight, T.M. Plant-Pollinator Interactions over 120 Years: Loss of Species, Co-Occurrence, and Function. Science 2013, 339, 1611–1615. [Google Scholar] [CrossRef] [PubMed]
- Allesina, S.; Pascual, M. Network structure, predator-prey modules, and stability in large food webs. Theor. Ecol. 2008, 1, 55–64. [Google Scholar] [CrossRef]
- Montesinos-Navarro, A.; Segarra-Moragues, J.G.; Valiente-Banuet, A.; Verdu, M. The network structure of plant-arbuscular mycorrhizal fungi. New Phytol. 2012, 194, 536–547. [Google Scholar] [CrossRef]
- Chagnon, P.L.; Bradley, R.L.; Klironomos, J.N. Using ecological network theory to evaluate the causes and consequences of arbuscular mycorrhizal community structure. New Phytol. 2012, 194, 307–312. [Google Scholar] [CrossRef]
- Tylianakis, J.M.; Martinez-Garcia, L.B.; Richardson, S.J.; Peltzer, D.A.; Dickie, I.A. Symmetric assembly and disassembly processes in an ecological network. Ecol. Lett. 2018, 21, 896–904. [Google Scholar] [CrossRef]
- Hacquard, S. Disentangling the factors shaping microbiota composition across the plant holobiont. New Phytol. 2016, 209, 454–457. [Google Scholar] [CrossRef]
- Derocles, S.A.P.; Bohan, D.A.; Dumbrell, A.J.; Kitson, J.J.N.; Massol, F.; Pauvert, C.; Plantegenest, M.; Vacher, C.; Evans, D.M. Chapter one—Biomonitoring for the 21st century: Integrating next-generation sequencing into ecological network analysis. In Advances in Ecological Research; Bohan, D.A., Dumbrell, A.J., Woodward, G., Jackson, M., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 58, pp. 1–62. [Google Scholar]
- Bennett, A.E.; Evans, D.M.; Powell, J.R. Potentials and pitfalls in the analysis of bipartite networks to understand plant-microbe interactions in changing environments. Funct. Ecol. 2019, 33, 107–117. [Google Scholar] [CrossRef]
- Tylianakis, J.M.; Tscharntke, T.; Lewis, O.T. Habitat modification alters the structure of tropical host–parasitoid food webs. Nature 2007, 445, 202–205. [Google Scholar] [CrossRef]
- Poudel, R.; Meyer, L.; Jumpponen, A.; Kennelly, M.M.; Rivard, C.L.; Schlatter, D.C.; Paulitz, T.C.; Gardener, B.M.; Kinkel, L.L.; Garrett, K.A. Microbiome networks: A systems framework for identifying candidate microbial assemblages for disease management in the era of genomics and phytobiomes. Phytopathology 2017, 107, 154. [Google Scholar]
- Bennett, A.E.; Daniell, T.J.; Oepik, M.; Davison, J.; Moora, M.; Zobel, M.; Selosse, M.-A.; Evans, D. Arbuscular mycorrhizal fungal networks vary throughout the growing season and between successional stages. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Sato, H.; Yamamoto, S.; Tanabe, A.S. Structural diversity across arbuscular mycorrhizal, ectomycorrhizal, and endophytic plant-fungus networks. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Polme, S.; Bahram, M.; Jacquemyn, H.; Kennedy, P.; Kohout, P.; Moora, M.; Oja, J.; Opik, M.; Pecoraro, L.; Tedersoo, L. Host preference and network properties in biotrophic plant-fungal associations. New Phytol. 2018, 217, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Staab, M.; Blüthgen, N.; Klein, A.-M. Tree diversity alters the structure of a tri-trophic network in a biodiversity experiment. Oikos 2015, 124, 827–834. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Bruelheide, H.; Chen, X.F.; Eichenberg, D.; Krober, W.; Xu, W.X.; Xu, L.T.; Schuldt, A. Tree diversity promotes generalist herbivore community patterns in a young subtropical forest experiment. Oecologia 2017, 183, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.-X.; Klein, A.-M.; Zhu, C.; Staab, M.; Durka, W.; Fischer, M.; Fornoff, F. Intra- and interspecific tree diversity promotes multitrophic plant–hemiptera–ant interactions in a forest diversity experiment. Basic Appl. Ecol. 2018, 29, 89–97. [Google Scholar] [CrossRef]
- Fornoff, F.; Klein, A.-M.; Blüthgen, N.; Staab, M. Tree diversity increases robustness of multi-trophic interactions. Proc. Biol. Sci. 2019, 286, 20182399. [Google Scholar] [CrossRef]
- Bruelheide, H.; Nadrowski, K.; Assmann, T.; Bauhus, J.; Both, S.; Buscot, F.; Chen, X.Y.; Ding, B.Y.; Durka, W.; Erfmeier, A.; et al. Designing forest biodiversity experiments: General considerations illustrated by a new large experiment in subtropical china. Methods Ecol. Evol. 2014, 5, 74–89. [Google Scholar] [CrossRef]
- Bruelheide, H. The Role of Tree and Shrub Diversity for Production, Erosion Control, Element Cycling, and Species Conservation in Chinese Subtropical Forest Ecosystems; Proposal for the 2nd of the DFG Research Unit 891 phase (2011–2014) (DFG Forschergruppe 891); Institute of Biology/Geobotany and Botanical Garden Martin Luther University Halle-Wittenberg: Halle, Germany, 2010; pp. 1–578. [Google Scholar]
- Chytrý, M.; Tichý, L.; Holt, J.; Botta-Dukát, Z. Determination of diagnostic species with statistical fidelity measures. J. Veg. Sci. 2002, 13, 79–90. [Google Scholar] [CrossRef]
- Stone, L.; Roberts, A. The checkerboard score and species distributions. Oecologia 1990, 85, 74–79. [Google Scholar] [CrossRef]
- Fodor, E. Linking biodiversity to mutualistic networks-woody species and ectomycorrhizal fungi. Ann. For. Res. 2013, 56, 53–78. [Google Scholar]
- Guimarães, P.R.; Rico-Gray, V.; Oliveira, P.S.; Izzo, T.J.; dos Reis, S.F.; Thompson, J.N. Interaction intimacy affects structure and coevolutionary dynamics in mutualistic networks. Curr. Biol. 2007, 17, 1797–1803. [Google Scholar]
- Guimerà, R.; Nunes Amaral, L.A. Functional cartography of complex metabolic networks. Nature 2005, 433, 895. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Härdtle, W.; Bruelheide, H.; Nadrowski, K.; Scholten, T.; von Wehrden, H.; von Oheimb, G. Site and neighborhood effects on growth of tree saplings in subtropical plantations (china). For. Ecol. Manag. 2014, 327, 118–127. [Google Scholar] [CrossRef]
- Weißbecker, C.; Buscot, F.; Wubet, T. Preservation of nucleic acids by freeze-drying for next generation sequencing analyses of soil microbial communities. J. Plant Ecol. 2017, 10, 81–90. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Boyer, F.; Mercier, C.; Bonin, A.; Le Bras, Y.; Taberlet, P.; Coissac, E. Obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 176–182. [Google Scholar] [CrossRef]
- Gaspar, J.M.; Thomas, W.K. Flowclus: Efficiently filtering and denoising pyrosequenced amplicons. Bioinformatics 2015, 16, 105. [Google Scholar] [CrossRef][Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. Vsearch: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.W.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. Funguild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 2 January 2019).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.P.; Veach, A.M.; Rigdon-Huss, A.R.; Grond, K.; Lickteig, S.K.; Lothamer, K.; Oliver, A.K.; Jumpponen, A. Scraping the bottom of the barrel: Are rare high throughput sequences artifacts? Fungal Ecol. 2015, 13, 221–225. [Google Scholar] [CrossRef]
- Morgan, M. Biocmanager: Access the Bioconductor Project Package Repository. R package version 1.30.4. 2018. Available online: https://rdrr.io/cran/BiocManager/f/vignettes/BiocManager.Rmd (accessed on 2 January 2019).
- McMurdie, P.J.; Paulson, J.N. Biomformat: An Interface Package for the Biom File Format. R/bioconductor package version 1.0.0. 2015. Available online: https://bioconductor.org/packages/devel/bioc/vignettes/biomformat/inst/doc/biomformat.html (accessed on 2 January 2019).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. R package version 0.8.0.1. 2019. Available online: https://github.com/tidyverse/dplyr (accessed on 15 February 2019).
- Dowle, M.; Srinivasan, A. Data.Table: Extension of “data.Frame”. R package version 1.12.0. 2019. Available online: https://cran.r-project.org/web/packages/data.table/index.html (accessed on 15 January 2019).
- Chang, W. Extrafont: Tools for Using Fonts. R package version 0.17. 2014. Available online: https://cran.r-project.org/web/packages/extrafont/index.html (accessed on 1 January 2019).
- Warnes, G.R.; Bolker, B.; Gorjanc, G.; Grothendieck, G.; Korosec, A.; Lumley, T.; MacQueen, D.; Magnusson, A.; Rogers, J. Gdata: Various R Programming Tools for Data Manipulation. R package version 2.18.0. 2017. Available online: https://cran.r-project.org/web/packages/gdata/index.html (accessed on 2 January 2019).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Wickham, H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef]
- Gerds, T.A. Prodlim: Product-limit Estimation for Censored Event History Analysis. R package version 2018.04.18. 2018. Available online: https://cran.r-project.org/web/packages/prodlim/index.html (accessed on 2 January 2019).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. Vegan: Community Ecology Package. R software package. 2013. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 January 2019).
- Dormann, C.F.; Gruber, B.; Fruend, J. Introducing the bipartite package: Analysing ecological networks. R News 2008, 8, 8–11. [Google Scholar]
- Giraudoux, P. Pgirmess: Spatial analysis and data mining for field ecologists, R package version 1.6.9. 2018. Available online: https://cran.r-project.org/web/packages/pgirmess/index.html (accessed on 2 January 2019).
- Almeida-Neto, M.; Guimarães, P.; Guimarães, P.R., Jr.; Loyola, R.D.; Ulrich, W. A consistent metric for nestedness analysis in ecological systems: Reconciling concept and measurement. Oikos 2008, 117, 1227–1239. [Google Scholar] [CrossRef]
- Ihaka, R.; Murrell, P.; Hornik, K.; Fisher, J.C.; Stauffer, R.; Wilke, C.O.; McWhite, C.D.; Zeileis, A. Colorspace: A toolbox for manipulating and assessing colors and palettes. R package version 1.4-0. 2019. [Google Scholar]
- Galili, T. Dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Paquette, A.; Messier, C. The effect of biodiversity on tree productivity: From temperate to boreal forests. Glob. Ecol. Biogeogr. 2011, 20, 170–180. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, H.Y.H.; Reich, P.B. Forest productivity increases with evenness, species richness and trait variation: A global meta-analysis. J. Ecol. 2012, 100, 742–749. [Google Scholar] [CrossRef]
- Forrester, D.I.; Bauhus, J. A review of processes behind diversity-productivity relationships in forests. Curr. For. Rep. 2016, 2, 45–61. [Google Scholar] [CrossRef]
- Fichtner, A.; Härdtle, W.; Bruelheide, H.; Kunz, M.; Li, Y.; von Oheimb, G. Neighbourhood interactions drive overyielding in mixed-species tree communities. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Gonzalez, A.M.M.; Dalsgaard, B.; Olesen, J.M. Centrality measures and the importance of generalist species in pollination networks. Ecol. Complex. 2010, 7, 36–43. [Google Scholar] [CrossRef]
- Blüthgen, N.; Menzel, F.; Hovestadt, T.; Fiala, B.; Bluthgen, N. Specialization, constraints, and conflicting interests in mutualistic networks. Curr. Biol. 2007, 17, 341–346. [Google Scholar] [CrossRef]
- Sánchez, F.; del Río, M.; Cañellas, I. Using historic management records to characterize the effects of management on the structural diversity of forests. For. Ecol. Manag. 2005, 207, 279–293. [Google Scholar]
- Bachelot, B.; Uriarte, M.; Zimmerman, J.K.; Thompson, J.; Leff, J.W.; Asiaii, A.; Koshner, J.; McGuire, K. Long-lasting effects of land use history on soil fungal communities in second-growth tropical rain forests. Ecol. appl. 2016, 26, 1881–1895. [Google Scholar] [CrossRef]
- Buscot, F. Implication of evolution and diversity in arbuscular and ectomycorrhizal symbioses. J. Plant. Physiol. 2015, 172, 55–61. [Google Scholar] [CrossRef]
- Alexander, I.J.; Lee, S.S. Mycorrhizas and ecosystem processes in tropical rain forest: Implications for diversity. In Biotic Interactions in the Tropics: Their Role in the Maintenance of Species Diversity; Burslem, D., Pinard, M., Hartley, S., Eds.; Cambridge University Press: Cambridge, UK, 2005; pp. 165–203. [Google Scholar]
- McGuire, K.L.; Allison, S.D.; Fierer, N.; Treseder, K.K. Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons. PLoS ONE 2013, 8, e68278. [Google Scholar] [CrossRef]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 2nd ed.; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Öpik, M.; Davison, J.; Moora, M.; Zobel, M. DNA-based detection and identification of Glomeromycota: The virtual taxonomy of environmental sequences. Botany 2013, 92, 135–147. [Google Scholar] [CrossRef]
- Wubet, T.; Kottke, I.; Teketay, D.; Oberwinkler, F. Arbuscular mycorrhizal fungal community structures differ between co-occurring tree species of dry afromontane tropical forest, and their seedlings exhibit potential to trap isolates suited for reforestation. Mycol. Prog. 2009, 8, 317–328. [Google Scholar] [CrossRef]
- Öpik, M.; Metsis, M.; Daniell, T.J.; Zobel, M.; Moora, M. Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol. 2009, 184, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, L.B.; Richardson, S.J.; Tylianakis, J.M.; Peltzer, D.A.; Dickie, I.A. Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytol. 2015, 205, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Harend, H.; Tedersoo, L. Network perspectives of ectomycorrhizal associations. Fungal Ecol. 2014, 7, 70–77. [Google Scholar] [CrossRef]
- Sepp, S.K.; Davison, J.; Jairus, T.; Vasar, M.; Moora, M.; Zobel, M.; Opik, M. Non-random association patterns in a plant-mycorrhizal fungal network reveal host-symbiont specificity. Mol. Ecol. 2019, 28, 365–378. [Google Scholar] [CrossRef]
- Toju, H.; Guimaraes, P.R.; Olesen, J.M.; Thompson, J.N. Assembly of complex plant-fungus networks. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Bascompte, J.; Jordano, P.; Melián, C.J.; Olesen, J.M. The nested assembly of plant–animal mutualistic networks. Proc. Natl. Acad. Sci. USA 2003, 100, 9383–9387. [Google Scholar] [CrossRef]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, K.; Vanhellemont, M.; Auge, H.; Baeten, L.; Baraloto, C.; Barsoum, N.; Bilodeau-Gauthier, S.; Bruelheide, H.; Castagneyrol, B.; Godbold, D.; et al. Contributions of a global network of tree diversity experiments to sustainable forest plantations. Ambio 2016, 45, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Tuck, S.L.; O’Brien, M.J.; Philipson, C.D.; Saner, P.; Tanadini, M.; Dzulkifli, D.; Godfray, H.C.J.; Godoong, E.; Nilus, R.; Ong, R.C.; et al. The value of biodiversity for the functioning of tropical forests: Insurance effects during the first decade of the sabah biodiversity experiment. Proc. Biol. Sci. 2016, 283. [Google Scholar] [CrossRef] [PubMed]
- Weißbecker, C.; Wubet, T.; Lentendu, G.; Buscot, F. BEF China Fungal Diversity: Raw Sequence Data of 454 Fungal ITS Amplicons. European Nucleotide Archive, 2016. Available online: https://www.ebi.ac.uk/ena/data/view/PRJEB12020 (accessed on 10 October 2018).
- Weißbecker, C.; Wubet, T. BEF china Fungal Diversity: R Statistic Scripts. Zenodo Data Archive, Version 2. 2018. Available online: https://zenodo.org/record/1215505 (accessed on 10 October 2018).
- Weißbecker, C.; Heintz-Buschart, A. Scripts and Supplementary Data Sets. Zenodo Data Archive, Version 2. 2019. Available online: https://zenodo.org/record/3533732 (accessed on 9 November 2019).
- Bissinger, V.; Kolditz, O. Helmholtz Interdisciplinary Graduate School for Environmental Research (HIGRADE). GAIA-Ecol. Perspect. Sci. Soc. 2008, 17, 71–73. [Google Scholar] [CrossRef]
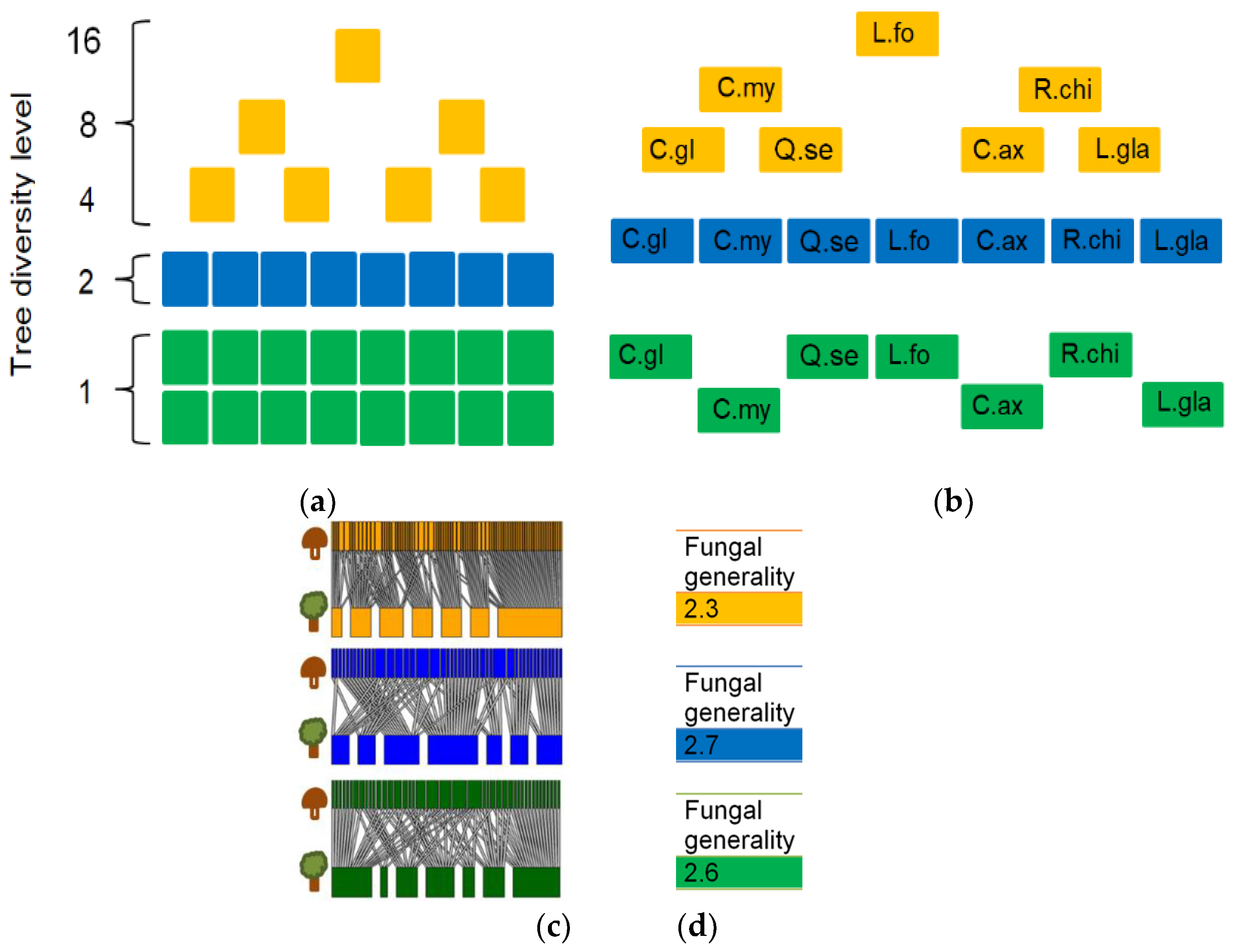
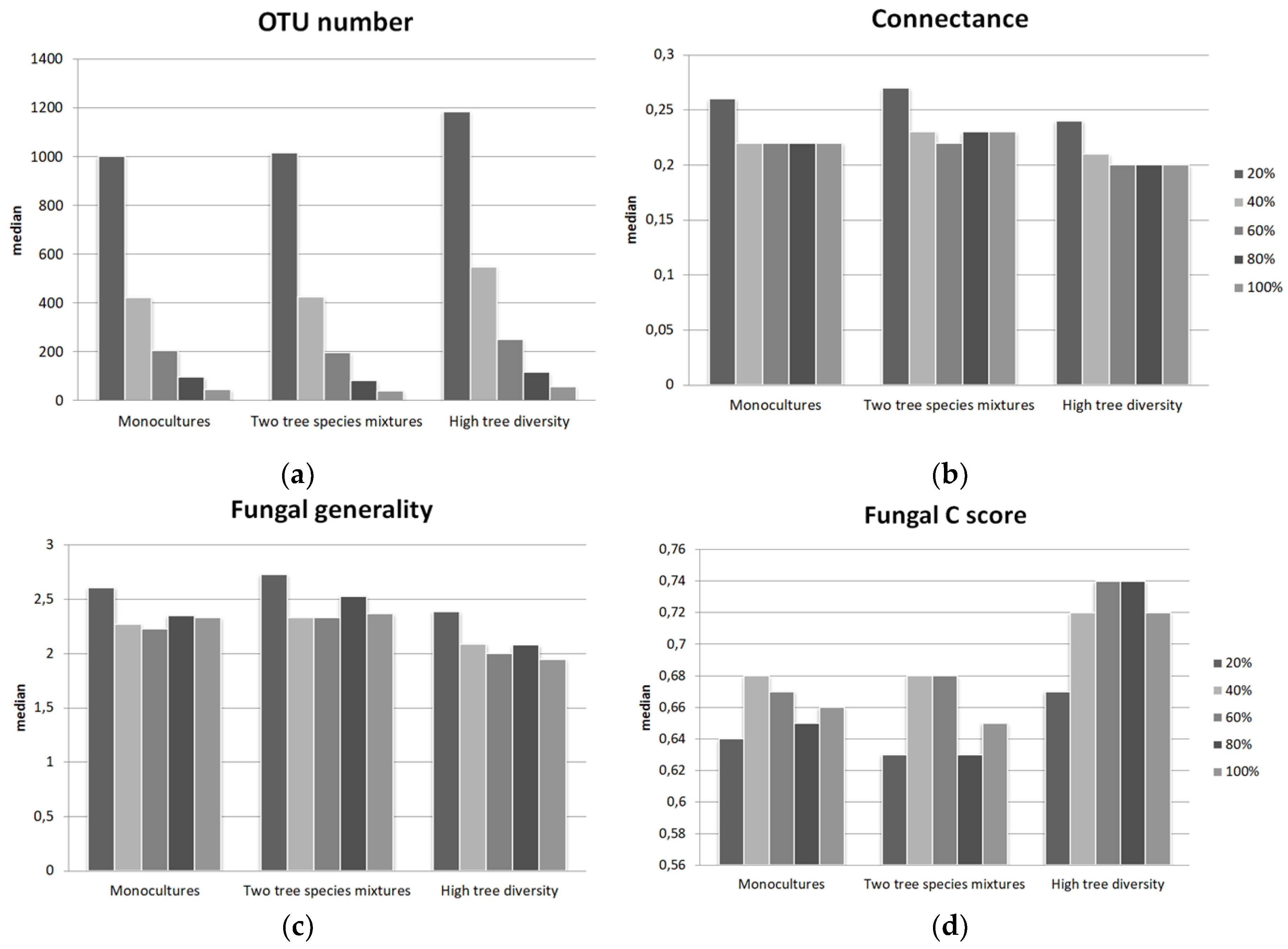

| Number of OTUs in Network | Modularity | Connectance 1 | Fungal Generality 2 | Fungal C Score 3 | Mean Number of Shared Fungal Partners 4 | Fungal OTU Richness | Fungal Shannon Diversity | |
|---|---|---|---|---|---|---|---|---|
| Kruskal p | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Median | ||||||||
| Mono | 206 | 0.52 | 0.22 | 2.26 | 0.67 | 9.43 | 1004 | 4.99 |
| Two mix. | 198 | 0.51 | 0.22 | 2.33 | 0.68 | 9.57 | 1017 | 5.06 |
| High | 251 | 0.58 | 0.2 | 2 | 0.74 | 8.48 | 1187 | 5.34 |
| Pairwise p | ||||||||
| 1-2 | <0.001 | n.s. | n.s. | <0.001 | n.s. | n.s. | n.s. | n.s. |
| 1-3 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| 2-3 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Networks | NODF Median | Wilcox.p |
|---|---|---|
| Tree monocultures | 21.51 | <0.001 |
| Null model | 57.6 | |
| Two tree species mixtures | 22.59 | <0.001 |
| Null model | 57.48 | |
| High tree species mixtures | 15.66 | <0.001 |
| Null model | 57.32 |
| This Study | [94] | [34] | [95] | [44] | [33] * | [26] * | [25] | |
|---|---|---|---|---|---|---|---|---|
| Study system | 16 subtropical tree species in a forest biodiversity experiment | Semi natural grasslands, 33 plant species | cool-temperate, warm-temperate and subtropical forests | Temperate forest with 33 tree species | Temperate forests, mainly Quercus and Carpinus | 33 understory plant species in temperate spruce forest | Xeric shrubland | |
| Country | China | Estonia | Japan | Japan | Romania | Estonia | Mexico | |
| Age | 3 years | 55–100 years | 25 years and 130 years | 130 years | ||||
| Treatment | Tree species diversity | Host plant functional group | Latitudinal gradient | Succession and seasonality | ||||
| Samples | Soil within tree rooting zone | Root samples | Root samples | Root samples | aboveground EcM fructifications | Root samples | Root samples | |
| Study target | Soil fungi | AM fungi | Soil fungi, fungal groups | Soil fungi | EcM fungi | AM fungi | AM fungi | |
| Nestedness | Less nested (15.66–29.42,) than random (53.87–60.04) NODF | More nested than random (27) nestedness temperature) | Anti-nested (−9 to −4) weighted NODF) | Less nested (25–35,) than random (32–40) weighted NODF | More nested (16) than random (38, 31) nestedness temperature) | More nested (14.36–54.83) than random, NODF | ||
| Number of modules | 1 | 5 | 8 | 4 | 5-9 | |||
| Modularity | 0.41–0.58 | Higher than random 0.18 | Moderate to low modularity (0.35–0.42), higher than random (0.32–0.38) | Low modularity 0.24 | 0.3–0.44 | Modular 0.30–0.57 | ||
| Connectance | 0.20–0.27 | Less connected than random 0.52 | 0.07 | 0.1-0.55 | High connectance 0.42 | Low connectance 0.05–0.15 | ||
| Fungal generality | 1.95–2.73 | 2.25–4.0 | ||||||
| Fungal C score | 0.63–0.74 | No difference of observed (0.59) and random value (0.58) | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weißbecker, C.; Heintz-Buschart, A.; Bruelheide, H.; Buscot, F.; Wubet, T. Linking Soil Fungal Generality to Tree Richness in Young Subtropical Chinese Forests. Microorganisms 2019, 7, 547. https://doi.org/10.3390/microorganisms7110547
Weißbecker C, Heintz-Buschart A, Bruelheide H, Buscot F, Wubet T. Linking Soil Fungal Generality to Tree Richness in Young Subtropical Chinese Forests. Microorganisms. 2019; 7(11):547. https://doi.org/10.3390/microorganisms7110547
Chicago/Turabian StyleWeißbecker, Christina, Anna Heintz-Buschart, Helge Bruelheide, François Buscot, and Tesfaye Wubet. 2019. "Linking Soil Fungal Generality to Tree Richness in Young Subtropical Chinese Forests" Microorganisms 7, no. 11: 547. https://doi.org/10.3390/microorganisms7110547
APA StyleWeißbecker, C., Heintz-Buschart, A., Bruelheide, H., Buscot, F., & Wubet, T. (2019). Linking Soil Fungal Generality to Tree Richness in Young Subtropical Chinese Forests. Microorganisms, 7(11), 547. https://doi.org/10.3390/microorganisms7110547






