NK Cells from RAG- or DCLRE1C-Deficient Patients Inhibit HCMV
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Cells
2.2. Preparation of Viral Stocks and Focal Expansion Assay
2.3. Flow Cytometry and IFN-γ Detection
2.4. Statistics
3. Results
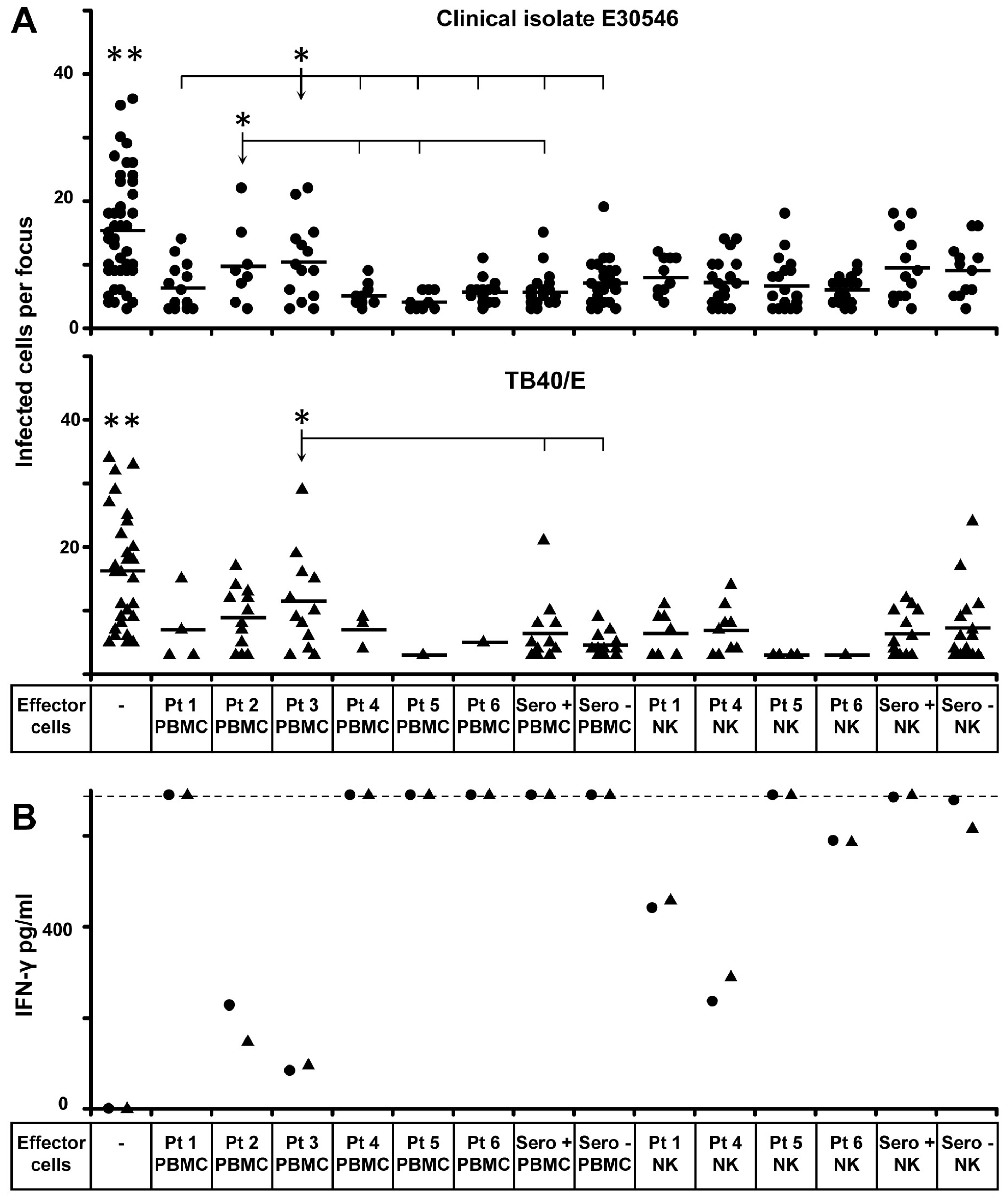
3.1. Inhibition of HCMV Transmission by NK Cells from SCID Patients with Defective RAGs or DCLRE1C (RAGs−/DCLRE1C−-NK)
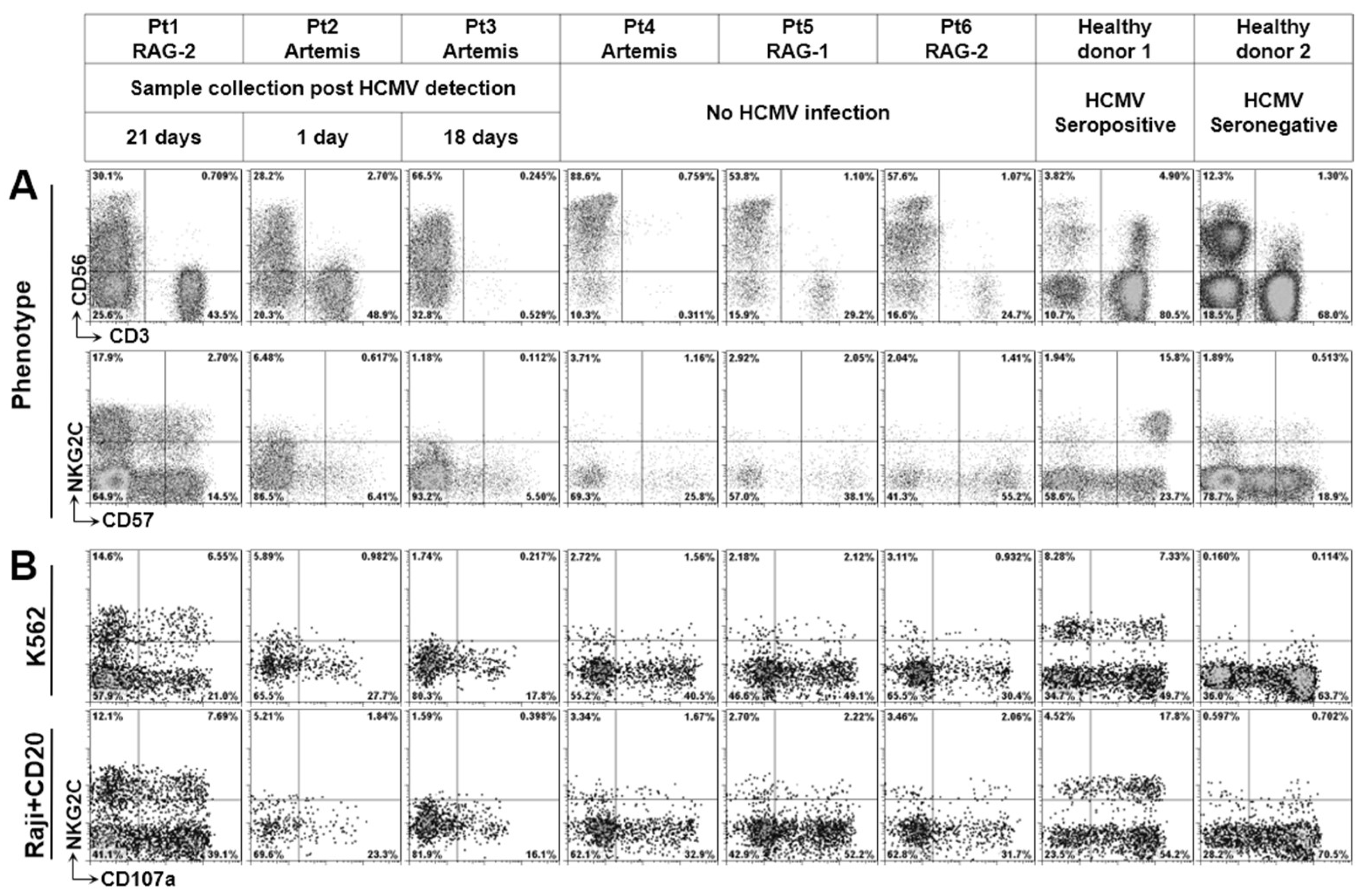
3.2. Phenotype of NK Cells from Defective V(D)J Recombination SCID
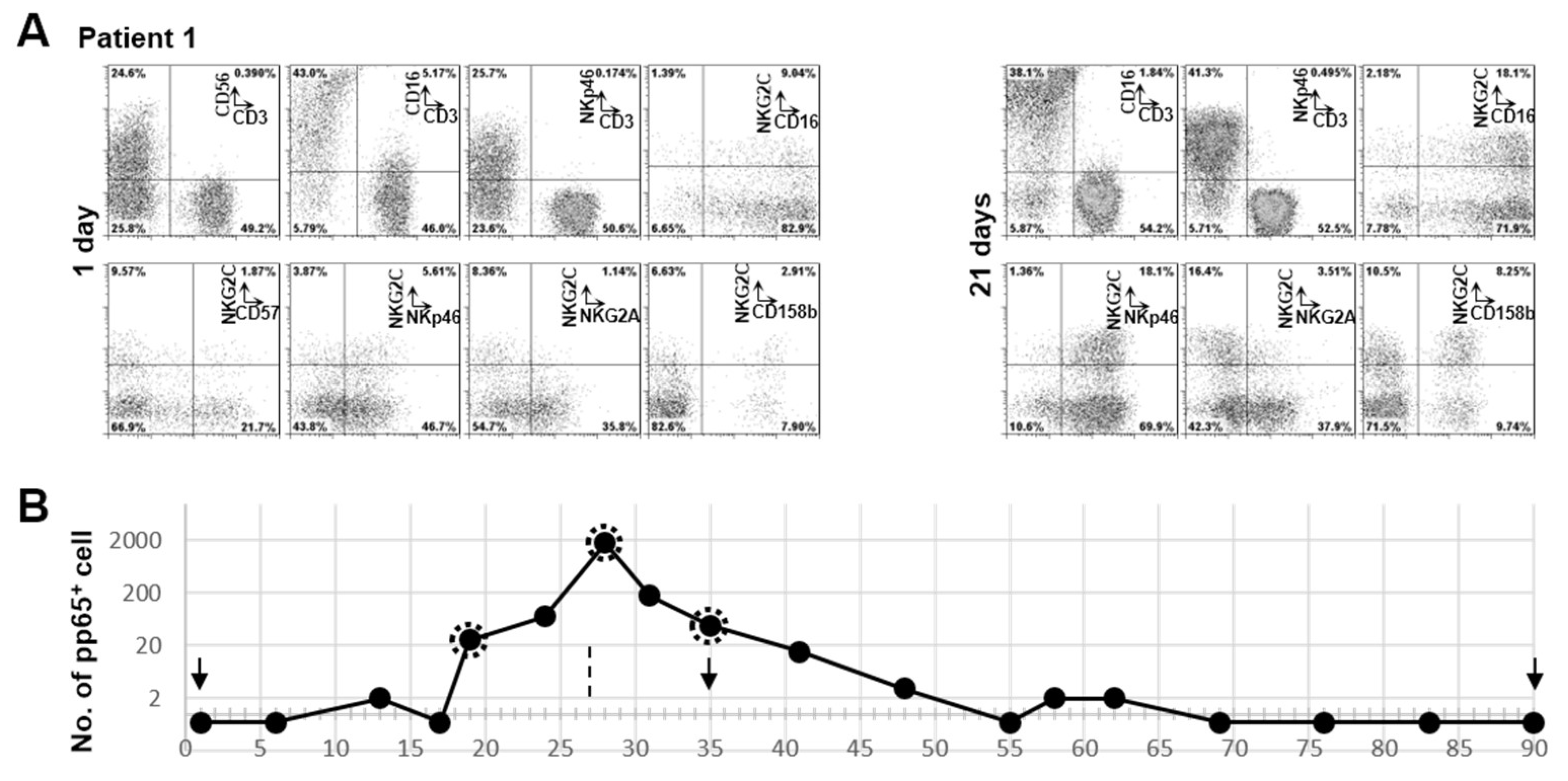
3.3. Clonal Expansion of NKG2C+ NK Cell in Patient 1 with RAG-2 Deficiency Post HCMV Acute Infection
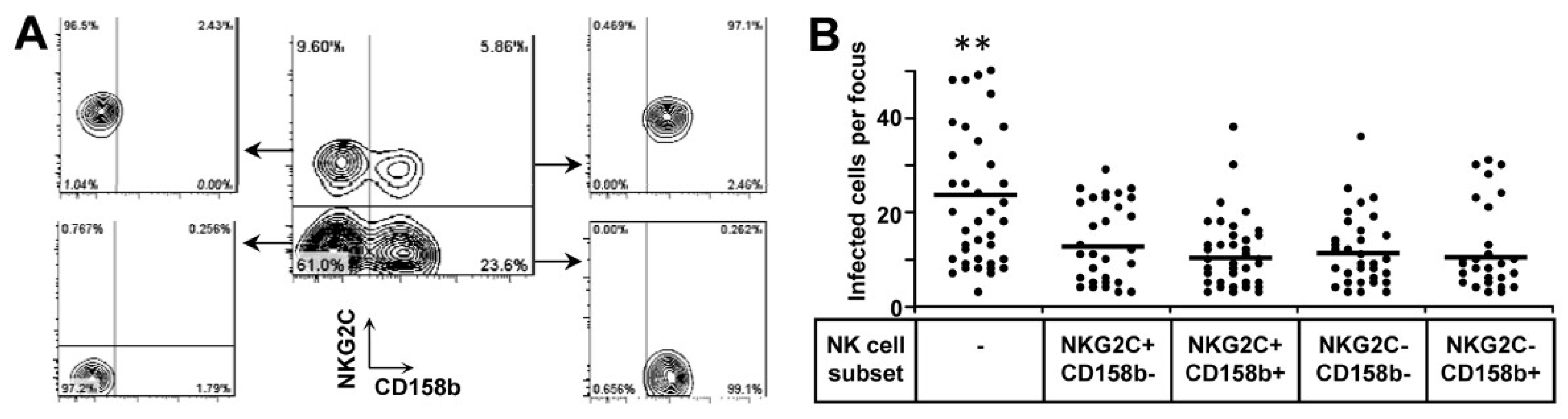
3.4. Both NKG2C+ and NKG2C− NK Cells Are Functional to Inhibit HCMV Transmission
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Britt, W. Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 2008, 325, 417–470. [Google Scholar] [PubMed]
- Schatz, D.G.; Ji, Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011, 11, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Malu, S.; Malshetty, V.; Francis, D.; Cortes, P. Role of non-homologous end joining in V(D)J recombination. Immunol. Res. 2012, 54, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- van der Burg, M.; van Veelen, L.R.; Verkaik, N.S.; Wiegant, W.W.; Hartwig, N.G.; Barendregt, B.H.; Brugmans, L.; Raams, A.; Jaspers, N.G.; Zdzienicka, M.Z.; et al. A new type of radiosensitive T-B-NK+ severe combined immunodeficiency caused by a LIG4 mutation. J. Clin. Investig. 2006, 116, 137–145. [Google Scholar] [CrossRef]
- Cowan, M.J.; Neven, B.; Cavazanna-Calvo, M.; Fischer, A.; Puck, J. Hematopoietic stem cell transplantation for severe combined immunodeficiency diseases. Biol. Blood Marrow Transplant. 2008, 14, 73–75. [Google Scholar] [CrossRef][Green Version]
- Schwarz, K.; Gauss, G.H.; Ludwig, L.; Pannicke, U.; Li, Z.; Lindner, D.; Friedrich, W.; Seger, R.A.; Hansen-Hagge, T.E.; Desiderio, S.; et al. RAG mutations in human B cell-negative SCID. Science 1996, 274, 97–99. [Google Scholar] [CrossRef]
- Vivier, E.; Raulet, D.H.; Moretta, A.; Caligiuri, M.A.; Zitvogel, L.; Lanier, L.L.; Yokoyama, W.M.; Ugolini, S. Innate or adaptive immunity? The example of natural killer cells. Science 2011, 331, 44–49. [Google Scholar] [CrossRef]
- Karo, J.M.; Schatz, D.G.; Sun, J.C. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell 2014, 159, 94–107. [Google Scholar] [CrossRef]
- Biron, C.A.; Byron, K.S.; Sullivan, J.L. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 1989, 320, 1731–1735. [Google Scholar] [CrossRef]
- Wu, Z.; Sinzger, C.; Reichel, J.J.; Just, M.; Mertens, T. Natural killer cells can inhibit the transmission of human cytomegalovirus in cell culture by using mechanisms from innate and adaptive immune responses. J. Virol. 2015, 89, 2906–2917. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sinzger, C.; Frascaroli, G.; Reichel, J.; Bayer, C.; Wang, L.; Schirmbeck, R.; Mertens, T. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J. Virol. 2013, 87, 7717–7725. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Frascaroli, G.; Bayer, C.; Schmal, T.; Mertens, T. Interleukin-2 from Adaptive T Cells Enhances Natural Killer Cell Activity against Human Cytomegalovirus-Infected Macrophages. J. Virol. 2015, 89, 6435–6441. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Wu, Z.; Reister, F.; Sampaio, K.L.; Frascaroli, G.; Cicin-Sain, L.; Mertens, T. Naive T cells are activated by autologous HCMV-infected endothelial cells through NKG2D and can control HCMV transmission in vitro. J. Gen. Virol. 2017, 98, 3068–3085. [Google Scholar] [CrossRef] [PubMed]
- Kutukculer, N.; Gulez, N.; Karaca, N.E.; Aksu, G.; Berdeli, A. Novel mutations and diverse clinical phenotypes in recombinase-activating gene 1 deficiency. Ital. J. Pediatr. 2012, 38, 8. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.M.; Ege, M.; Pottharst, A.; Schulz, A.S.; Schwarz, K.; Friedrich, W. Transplacentally acquired maternal T lymphocytes in severe combined immunodeficiency: A study of 121 patients. Blood 2001, 98, 1847–1851. [Google Scholar] [CrossRef]
- Comans-Bitter, W.M.; de Groot, R.; van den Beemd, R.; Neijens, H.J.; Hop, W.C.; Groeneveld, K.; Hooijkaas, H.; van Dongen, J.J. Immunophenotyping of blood lymphocytes in childhood. Reference values for lymphocyte subpopulations. J. Pediatr. 1997, 130, 388–393. [Google Scholar] [CrossRef]
- Ivarsson, M.A.; Loh, L.; Marquardt, N.; Kekäläinen, E.; Berglin, L.; Björkström, N.K.; Westgren, M.; Nixon, D.F.; Michaëlsson, J. Differentiation and functional regulation of human fetal NK cells. J. Clin. Investig. 2013, 123, 3889–3901. [Google Scholar] [CrossRef]
- Dalle, J.H.; Menezes, J.; Wagner, E.; Blagdon, M.; Champagne, J.; Champagne, M.A.; Duval, M. Characterization of cord blood natural killer cells: Implications for transplantation and neonatal infections. Pediatr. Res. 2005, 57, 649–655. [Google Scholar] [CrossRef]
- Beziat, V.; Liu, L.L.; Malmberg, J.A.; Ivarsson, M.A.; Sohlberg, E.; Björklund, A.T.; Retière, C.; Sverremark-Ekström, E.; Traherne, J.; Ljungman, P.; et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013, 121, 2678–2688. [Google Scholar] [CrossRef]
- Zambello, R.; Teramo, A.; Barila, G.; Gattazzo, C.; Semenzato, G. Activating KIRs in Chronic Lymphoproliferative Disorder of NK Cells: Protection from Viruses and Disease Induction? Front. Immunol. 2014, 5, 72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guma, M.; Angulo, A.; Vilches, C.; Gomez-Lozano, N.; Malats, N.; Lopez-Botet, M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Scott, J.M.; Hwang, I.; Kim, S. Cutting edge: Antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 2013, 190, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Landskron, J.; Ask, E.H.; Enqvist, M.; Sohlberg, E.; Traherne, J.A.; Hammer, Q.; Goodridge, J.P.; Larsson, S.; Jayaraman, J.; et al. Critical Role of CD2 Co-stimulation in Adaptive Natural Killer Cell Responses Revealed in NKG2C-Deficient Humans. Cell Rep. 2016, 15, 1088–1099. [Google Scholar] [CrossRef]
- Tomasec, P.; Braud, V.M.; Rickards, C.; Powell, M.B.; McSharry, B.P.; Gadola, S.; Cerundolo, V.; Borysiewicz, L.K.; McMichael, A.J.; Wilkinson, G.W. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000, 287, 1031. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Magri, G.; Muntasell, A.; Romo, N.; Sáez-Borderías, A.; Pende, D.; Geraghty, D.E.; Hengel, H.; Angulo, A.; Moretta, A.; López-Botet, M. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood 2011, 117, 848–856. [Google Scholar] [CrossRef]
- Dobbs, K.; Tabellini, G.; Calzoni, E.; Patrizi, O.; Martinez, P.; Giliani, S.C.; Moratto, D.; Al-Herz, W.; Cancrini, C.; Cowan, M.; et al. Natural Killer Cells from Patients with Recombinase-Activating Gene and Non-Homologous End Joining Gene Defects Comprise a Higher Frequency of CD56(bright) NKG2A (+++) Cells, and Yet Display Increased Degranulation and Higher Perforin Content. Front. Immunol. 2017, 8, 798. [Google Scholar] [CrossRef]
- Bigley, A.B.; Spielmann, G.; Agha, N.; O’Connor, D.P.; Simpson, R.J. Dichotomous effects of latent CMV infection on the phenotype and functional properties of CD8+ T-cells and NK-cells. Cell. Immunol. 2016, 300, 26–32. [Google Scholar] [CrossRef]
- Kuijpers, T.W.; Baars, P.A.; Dantin, C.; van den Burg, M.; van Lier, R.A.; Roosnek, E. Human NK cells can control CMV infection in the absence of T cells. Blood 2008, 112, 914–915. [Google Scholar] [CrossRef]
- Farnault, L.; Chambost, H.; Michel, G.; Thuret, I.; de Saint Basile, G.; Fischer, A.; Picard, C.; Picard, C.; Orlanducci, F.; Farnarier, C.; et al. Persistence of natural killer cells with expansion of a hypofunctional CD56-CD16+KIR+NKG2C+ subset in a patient with atypical Janus kinase 3-deficient severe combined immunodeficiency. J. Allergy Clin. Immunol. 2013, 131, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Beziat, V.; Sleiman, M.; Goodridge, J.P.; Kaarbo, M.; Liu, L.L.; Rollag, H.; Ljunggren, H.G.; Zimmer, J.; Malmberg, K.J. Polyclonal Expansion of NKG2C (+) NK Cells in TAP-Deficient Patients. Front. Immunol. 2015, 6, 507. [Google Scholar] [CrossRef] [PubMed]
- Rettman, P.; Willem, C.; David, G.; Riou, R.; Legrand, N.; Esbelin, J.; Cesbron, A.; Senitzer, D.; Gagne, K.; Retière, C. New insights on the natural killer cell repertoire from a thorough analysis of cord blood cells. J. Leukoc. Biol. 2016, 100, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.C.; Stanton, R.J.; Banat, J.J.; Wills, M.R. Leukocyte Immunoglobulin-Like Receptor 1-Expressing Human Natural Killer Cell Subsets Differentially Recognize Isolates of Human Cytomegalovirus through the Viral Major Histocompatibility Complex Class I Homolog UL18. J. Virol. 2016, 90, 3123–3137. [Google Scholar] [CrossRef] [PubMed]
- Rolle, A.; Meyer, M.; Calderazzo, S.; Jager, D.; Momburg, F. Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep. 2018, 24, 1967–1976 e4. [Google Scholar] [CrossRef]
- Hammer, Q.; Ruckert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef]
- Kaiser, B.K.; Pizarro, J.C.; Kerns, J.; Strong, R.K. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc. Natl. Acad. Sci. USA 2008, 105, 6696–6701. [Google Scholar] [CrossRef]
- Willcox, B.E.; Thomas, L.M.; Bjorkman, P.J. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 2003, 4, 913–919. [Google Scholar] [CrossRef]
- Yu, J.; Heller, G.; Chewning, J.; Kim, S.; Yokoyama, W.M.; Hsu, K.C. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 2007, 179, 5977–5989. [Google Scholar] [CrossRef]
- Leong, C.C.; Chapman, T.L.; Bjorkman, P.J.; Formankova, D.; Mocarski, E.S.; Phillips, J.H.; Lanier, L.L. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: The role of endogenous class I major histocompatibility complex and a viral class I homolog. J. Exp. Med. 1998, 187, 1681–1687. [Google Scholar] [CrossRef]
- Yu, K.; Davidson, C.L.; Wojtowicz, A.; Lisboa, L.; Wang, T.; Airo, A.M.; Villard, J.; Buratto, J.; Sandalova, T.; Achour, A.; et al. LILRB1 polymorphisms influence posttransplant HCMV susceptibility and ligand interactions. J. Clin. Investig. 2018, 128, 1523–1537. [Google Scholar] [CrossRef] [PubMed]
| Gender (f/m) | Known Mutations | Cell Count/μL | Age # | ||||
|---|---|---|---|---|---|---|---|
| T | B | NK | Virus Detection | ||||
| Pt 1 | f | RAG2: c.(572C>A); (572C>A), p.(Ser194X); (Ser194X) | 1300 * | 0 | 910 | 110 | VI **from urine |
| Pt 2 | m | Artemis: c.(1147C>T); (1147C>T), p.(Arg383X); (Arg383X) | 283 * | 0 | 554 | 122 | VI ** from urine |
| Pt 3 | m | Artemis: c.((?_-38)_246+?del); ((?_-38)_246+?del), p.(0);(0) | 3 | 0 | 306 | 96 | pp65 antigenemia |
| Pt 4 | f | Artemis: c.((?_-38)_246+?del); ((?_-38)_246+?del), p.(0);(0) | 0 | 0 | 1984 | 196 | - |
| Pt 5 | m | RAG1: c.(1331C>T); (1331C>T), p.(Ala444Val); (Ala444Val) | 392 | 0 | 1094 | 285 | - |
| Pt 6 | m | RAG2: c.(475C>T); (475C>T), p.(Arg159Cys); (Arg159Cys) | 198 | 0 | 276 | 199 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Subramanian, N.; Jacobsen, E.-M.; Laib Sampaio, K.; van der Merwe, J.; Hönig, M.; Mertens, T. NK Cells from RAG- or DCLRE1C-Deficient Patients Inhibit HCMV. Microorganisms 2019, 7, 546. https://doi.org/10.3390/microorganisms7110546
Wu Z, Subramanian N, Jacobsen E-M, Laib Sampaio K, van der Merwe J, Hönig M, Mertens T. NK Cells from RAG- or DCLRE1C-Deficient Patients Inhibit HCMV. Microorganisms. 2019; 7(11):546. https://doi.org/10.3390/microorganisms7110546
Chicago/Turabian StyleWu, Zeguang, Narmadha Subramanian, Eva-Maria Jacobsen, Kerstin Laib Sampaio, Johannes van der Merwe, Manfred Hönig, and Thomas Mertens. 2019. "NK Cells from RAG- or DCLRE1C-Deficient Patients Inhibit HCMV" Microorganisms 7, no. 11: 546. https://doi.org/10.3390/microorganisms7110546
APA StyleWu, Z., Subramanian, N., Jacobsen, E.-M., Laib Sampaio, K., van der Merwe, J., Hönig, M., & Mertens, T. (2019). NK Cells from RAG- or DCLRE1C-Deficient Patients Inhibit HCMV. Microorganisms, 7(11), 546. https://doi.org/10.3390/microorganisms7110546




