Integrated Process for Bioenergy Production and Water Recycling in the Dairy Industry: Selection of Kluyveromyces Strains for Direct Conversion of Concentrated Lactose-Rich Streams into Bioethanol
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source and Characterization of Nanofiltration (NF) and Reverse Osmosis (RO) Permeates and Retentates
2.2. Yeast Strains and Fermentation Assays
3. Results and Discussion
3.1. Chemical Composition of Nanofiltration and Reverse Osmosis Permeates and Retentates from Dairy Wastewater
3.1.1. Composition of NF and RO Permeates and Comparison with Drinking Water Regulation
3.1.2. Composition of the NF and RO Retentates
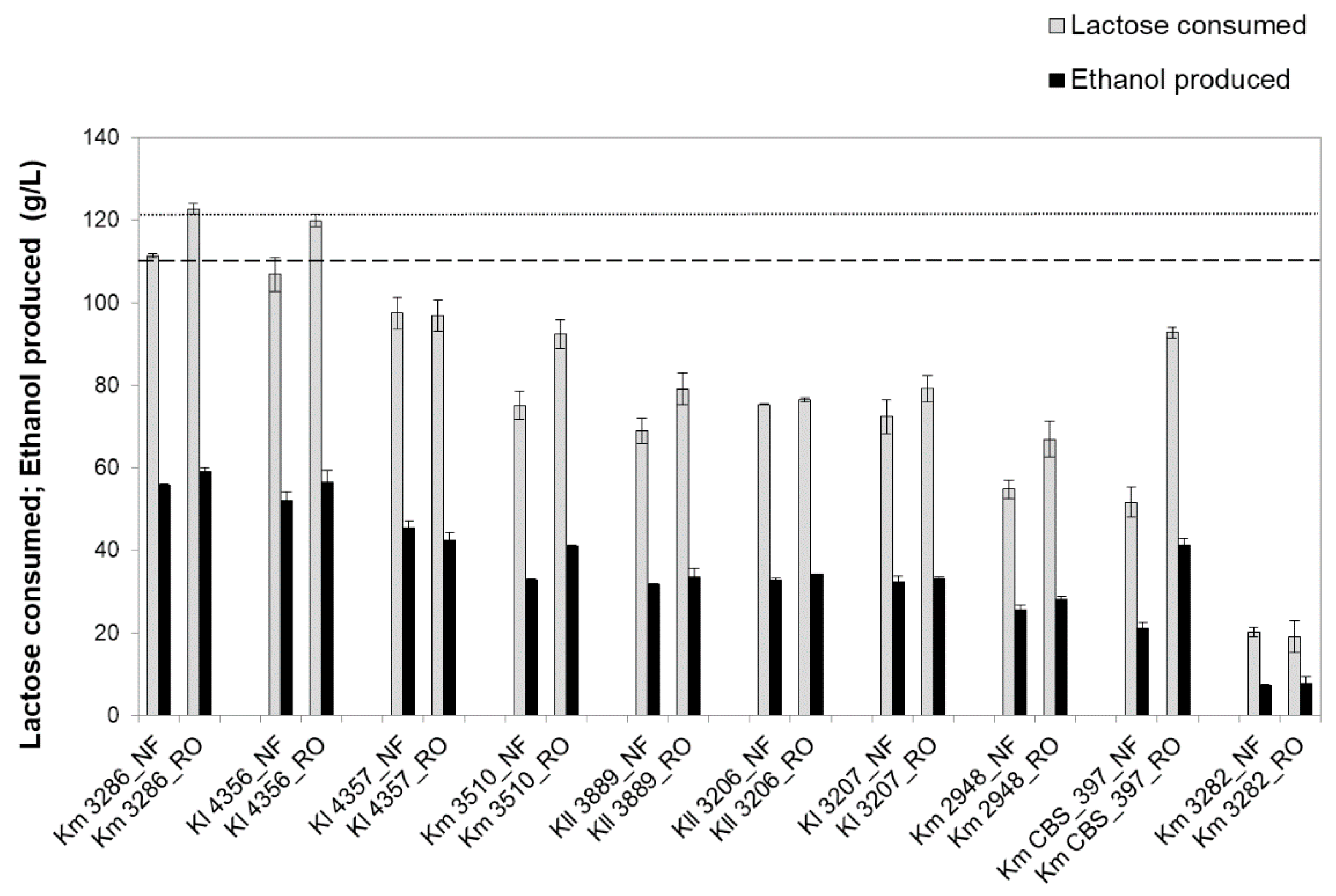
3.2. Screening of Kluyveromyces Strains for NF or RO Retentate Fermentation
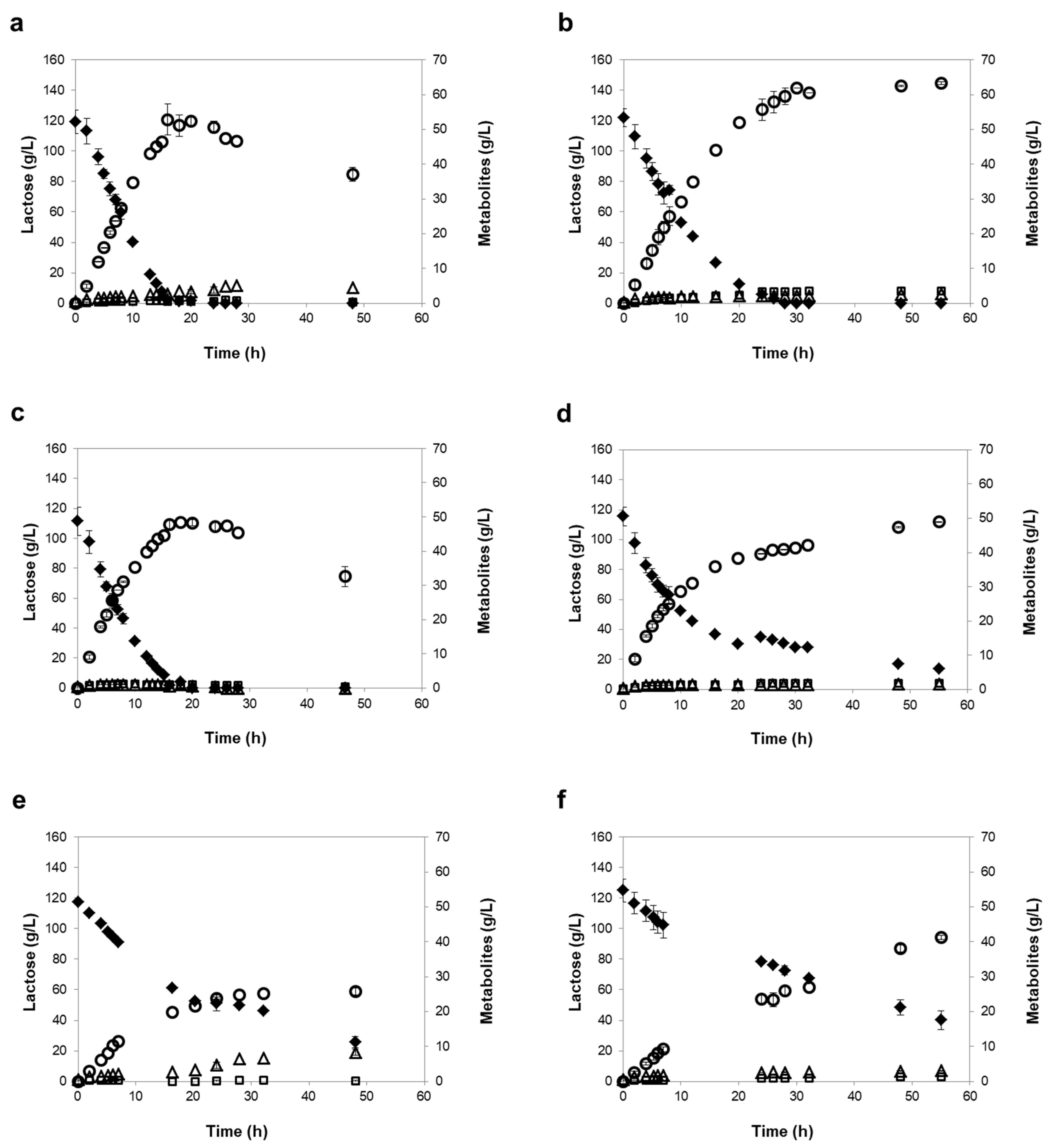
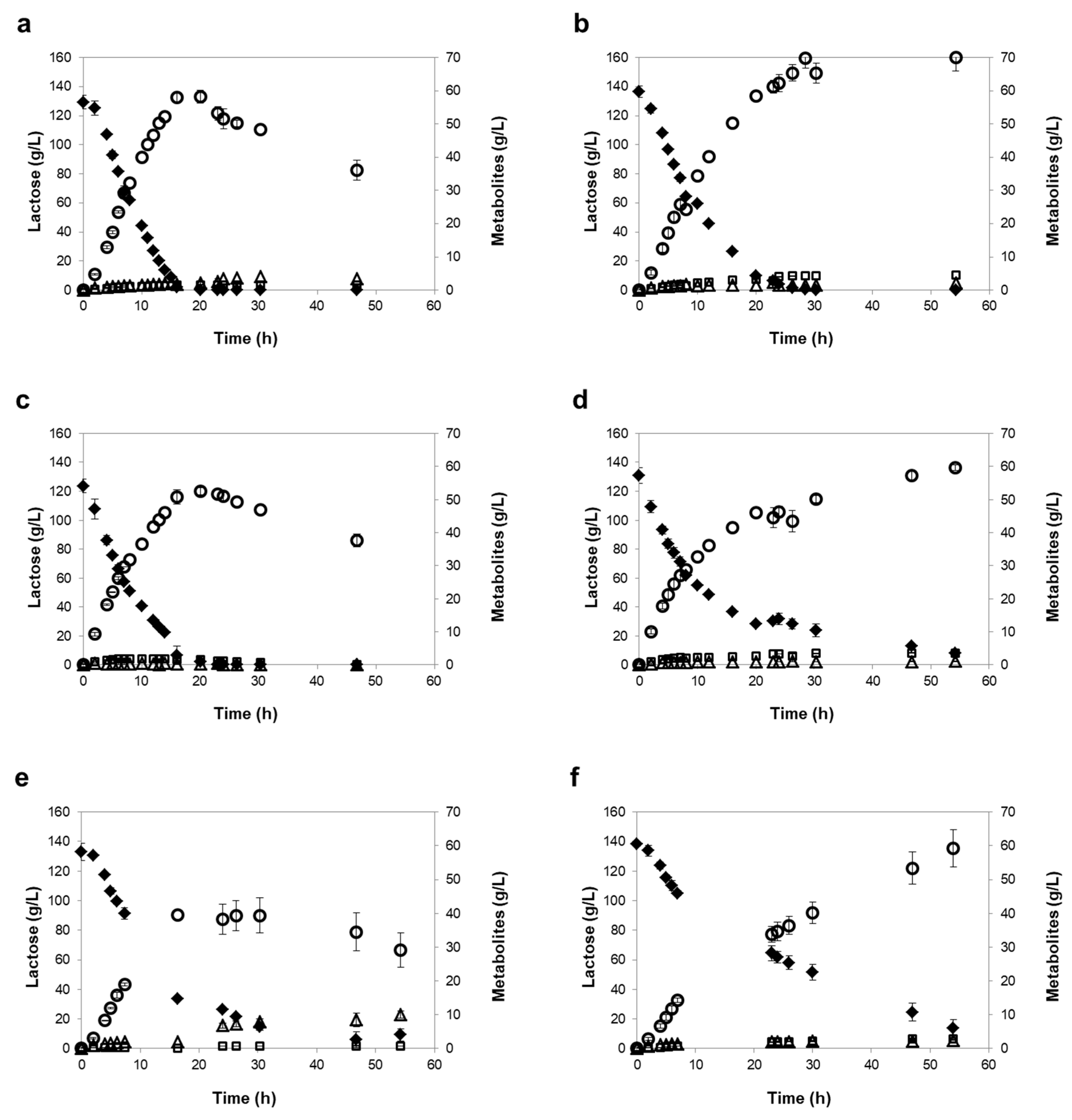
3.3. Influence of Different Oxygen-Limiting Conditions in NF or RO Rententate Fermentation Performance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Commission. U AGRICULTURAL OUTLOOK for the Agricultural Markets and Income 2017–2030. Technical Report. 2017. Available online: https://ec.europa.eu/info/sites/info/files/food-farming-fisheries/farming/documents/agricultural-outlook-2017-30_en.pdf (accessed on 8 October 2019).
- Britz, T.J.; Van Sckalkwyk, C.; Hung, Y.T. Treatment of dairy processing wastewater. In Handbook of Industrial and Hazardous Waste Treatment, 2nd ed.; Marcel Dekker Inc.: New York, NY, USA, 2004; pp. 616–646. [Google Scholar]
- Rad, S.J.; Lewis, M.J. Water utilisation, energy utilisation and waste water management in the dairy industry: A review. Int. J. Dairy Technol. 2014, 67, 1–20. [Google Scholar] [CrossRef]
- Dresch, M.; Daufin, G.; Chaufer, B. Membrane processes for the recovery of dairy cleaning-in-place solutions. Lait 1999, 79, 245–259. [Google Scholar] [CrossRef] [Green Version]
- Shahmansouri, A.; Bellona, C. Nanofiltration technology in water treatment and reuse: Applications and costs. Water Sci. Technol. 2015, 71, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Beaton, N.B. Ultrafiltration and reverse osmosis in the dairy industry-an introduction to sanitary considerations. J. Food Prot. 1979, 42, 584–590. [Google Scholar] [CrossRef]
- Park, H.-G.; Kwon, Y.-N. Long-term stability of low-pressure reverse osmosis (RO) membrane operation—A pilot scale study. Water 2018, 10, 93. [Google Scholar] [CrossRef]
- Early, R. Dairy products and milk-based food ingredients. In Natural Food Additives, Ingredients and Flavourings; Baines, D., Seal, R., Eds.; Woodhead Publishing Limited: Sawston, UK, 2012; pp. 417–445. [Google Scholar] [CrossRef]
- Meneses, Y.E.; Flores, R.A. Feasibility, safety, and economic implications of whey-recovered water in cleaning-in-place systems: A case study on water conservation for the dairy industry. J. Dairy Sci. 2016, 99, 3396–3407. [Google Scholar] [CrossRef] [Green Version]
- Domingues, L.; Guimarães, P.M.R.; Oliveira, C. Metabolic engineering of Saccharomyces cerevisiae for lactose/whey fermentation. Bioeng. Bugs 2010, 1, 164–171. [Google Scholar] [CrossRef]
- Guimarães, P.M.R.; Teixeira, J.A.; Domingues, L. Fermentation of lactose to bio-ethanol by yeasts as part of integrated solutions for the valorisation of cheese whey. Biotechnol. Adv. 2010, 28, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Hagman, A.; Säll, T.; Compagno, C.; Piskur, J. Yeast “Make-accumulate-consume” life strategy evolved as a multi-atep process that predates the whole genome duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef]
- Castro, R.C.A.; Roberto, I.C. Selection of a thermotolerant Kluyveromyces marxianus strain with potential application for cellulosic ethanol production by simultaneous saccharification and fermentation. Appl. Biochem. Biotechnol. 2014, 172, 1553–1564. [Google Scholar] [CrossRef]
- Belém, M.A.F.; Lee, B.H. Production of bioingredients from Kluyveromyces marxianus grown on whey: An alternative. Crit. Rev. Food Sci. Nutr. 1998, 38, 565–598. [Google Scholar] [CrossRef] [PubMed]
- Radecka, D.; Mukherjee, V.; Mateo, R.Q.; Stojiljkovic, M.; Foulquié-Moreno, M.R.; Thevelein, J.M. Looking beyond Saccharomyces: The potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef]
- Galbe, M.; Sassner, P.; Wingren, A.; Zacchi, G. Process engineering economics of bioethanol production. Adv. Biochem. Eng. Biotechnol. 2007, 108, 303–327. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Guidelines for Drinking-Water Quality. Technical Report. 2017. Available online: https://apps.who.int/iris/bitstream/handle/10665/254637/9789241549950-eng.pdf?sequence=1 (accessed on 8 October 2019).
- European Commission (EC). Council Directive 98/83/EC of 3 November 1998 on the Quality of Water Intended for Human Consumption. Technical Report. 1998. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:31998L0083&from=EN (accessed on 8 October 2019).
- United States Environmental Protection Agency (EPA). Ground Water and Drinking Water-National Primary Drinking Water Regulations; Technical Report; 2017. Available online: https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations#Inorganic (accessed on 8 October 2019).
- Japan Water Works Association (JWWA). JSupply of Drinking Water with Clean and Safe-Water Quality Standards of Drinking Water. Technical Report. 2007. Available online: http://www.jwwa.or.jp/english/water_en/water-e07.html (accessed on 8 October 2019).
- Jensen, N.B.; Strucko, T.; Kildegaard, K.R.; David, F.; Maury, J.; Mortensen, U.H.; Forster, J.; Nielsen, J.; Borodina, I. EasyClone: Method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 238–248. [Google Scholar] [CrossRef]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; Van Dijken, J.P. Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992, 8, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Smith, K.D.; Davis, J.H.; Gordon, P.B.; Breaker, R.R.; Strobel, S.A. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 19018–19023. [Google Scholar] [CrossRef] [Green Version]
- Murguia, J.R.; Belles, J.M.; Serrano, R. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 1996, 271, 29029–29033. [Google Scholar] [CrossRef]
- Blomberg, A.; Adler, L. Tolerance of fungi to NaCl. Mycol. Ser. 1993, 10, 209–232. [Google Scholar]
- Rajakala, P.; Karthigai Selvi, P. The effect of pH, temperature and alkali metal ions on the hydrolsis of whey lactose catalysed by β -galactosidase from Kluyveromyces marxianus. Int. J. Dairy Sci. 2006, 1, 167–172. [Google Scholar] [CrossRef]
- Díez-Antolínez, R.; Hijosa-Valsero, M.; Paniagua-García, A.I.; Garita-Cambronero, J.; Gómez, X. Yeast screening and cell immobilization on inert supports for ethanol production from cheese whey permeate with high lactose loads. PLoS ONE 2018, 13, e0210002. [Google Scholar] [CrossRef]
- Gabardo, S.; Rech, R.; Rosa, C.A.; Ayub, M.A.Z. Dynamics of ethanol production from whey and whey permeate by immobilized strains of Kluyveromyces marxianus in batch and continuous bioreactors. Renew. Energy 2014, 69, 89–96. [Google Scholar] [CrossRef]
- Janssens, J.H.; Burris, N.; Woodward, A.; Bailey, R.B. Lipid-enhanced ethanol production by Kluyveromyces fragilis. Appl. Environ. Microbiol. 1983, 45, 598–602. [Google Scholar] [PubMed]
- Mahmoud, M.M.; Kosikowski, F.V. Alcohol and single cell protein production by Kluyveromyces in concentrated whey permeates with reduced ash. J. Dairy Sci. 1982, 65, 2082–2087. [Google Scholar] [CrossRef]
- Vijaikishore, P.; Karanth, N.G. Glycerol production by fermentation. Appl. Biochem. Biotechnol. 1984, 9, 243–253. [Google Scholar] [CrossRef]
- Silveira, W.B.; Passos, F.J.V.; Mantovani, H.C.; Passos, F.M.L. Ethanol production from cheese whey permeate by Kluyveromyces marxianus UFV-3: A flux analysis of oxido-reductive metabolism as a function of lactose concentration and oxygen levels. Enzym. Microb. Technol. 2005, 36, 930–936. [Google Scholar] [CrossRef]
- Basso, L.C.; De Amorim, H.V.; De Oliveira, A.J.; Lopes, M.L. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008, 8, 1155–1163. [Google Scholar] [CrossRef]
- Liu, C.-G.; Hao, X.-M.; Lin, Y.-H.; Bai, F.-W. Redox potential driven aeration during very-high-gravity ethanol fermentation by using flocculating yeast. Sci. Rep. 2016, 6, 25763. [Google Scholar] [CrossRef]
- González Siso, M.I.; Cerdán, E.M. Kluyveromyces lactis: A suitable yeast model to study cellular defense mechanisms against hypoxia-induced oxidative stress. Oxidative Med. Cell. Longev. 2012, 2012, 14. [Google Scholar] [CrossRef]
- Lane, M.M.; Burke, N.; Karreman, R.; Wolfe, K.H.; O’Byrne, C.P.; Morrissey, J.P. Physiological and metabolic diversity in the yeast Kluyveromyces marxianus. Antonie Van Leeuwenhoek 2011, 100, 507–519. [Google Scholar] [CrossRef]
- Varela, J.A.; Montini, N.; Scully, D.; Van der Ploeg, R.; Oreb, M.; Boles, E.; Hirota, J.; Akada, R.; Hoshida, H.; Morrissey, J.P. Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Ryu, Y.; Jang, H.; Lee, H. Enhancement of ethanol tolerance of lactose assimilating yeast strain by protoplast fusion. J. Microbiol. Biotechnol. 1991, 1, 151–156. [Google Scholar]
- Sansonetti, S.; Curcio, S.; Calabrò, V.; Iorio, G. Bio-ethanol production by fermentation of ricotta cheese whey as an effective alternative non-vegetable source. Biomass Bioenergy 2009, 33, 1687–1692. [Google Scholar] [CrossRef]
- Guimarães, P.M.R.; François, J.; Parrou, J.L.; Teixeira, J.A.; Domingues, L. Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl. Environ. Microbiol. 2008, 74, 1748. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.-A.; Wang, G.-H.; Chen, A.-L.; Li, Y.-F.; Liu, J.-P.; Li, Y.-Y.; Bolotin-Fukuhara, M.; Bao, W.-G. Gene responses to oxygen availability in Kluyveromyces lactis: An insight on the evolution of the oxygen-responding system in yeast. PLoS ONE 2009, 4, e7561. [Google Scholar] [CrossRef] [PubMed]
- Snoek, I.S.; Steensma, H.Y. Why does Kluyveromyces lactis not grow under anaerobic conditions? Comparison of essential anaerobic genes of Saccharomyces cerevisiae with the Kluyveromyces lactis genome. FEMS Yeast Res. 2006, 6, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Madeira-Jr, J.V.; Gombert, A.K. Towards high-temperature fuel ethanol production using Kluyveromyces marxianus: On the search for plug-in strains for the Brazilian sugarcane-based biorefinery. Biomass Bioenergy 2018, 119, 217–228. [Google Scholar] [CrossRef]
- Rollini, M.; Trinetta, V.; Musatti, A.; Manzoni, M. Influence of substrate on β-galactosidase production by Kluyveromyces strains. Ann. Microbiol. 2008, 58, 705. [Google Scholar] [CrossRef]
| Kluyveromyces Strains | Numbering in Other Strain Collections |
|---|---|
| K. lactis PYCC 3207 | CBS 844 |
| K. lactis PYCC 4356 | |
| K. lactis PYCC 4357 | |
| K. lactis var. lactis PYCC 3206 | CBS 845 |
| K. lactis var. lactis PYCC 3889 | CBS 4574; DBVPG 6030; NRRL Y-2236; UCD 70-2 |
| K. marxianus PYCC 2948 | CBS 2762; ATCC 16045; CCRC 21623; DBVPG 6071; NCYC 970; NRRL YB-4327; UCD 71-13; ISA 1034 |
| K. marxianus PYCC 3282 | CBS 608; NCYC 143; NCTC 1302 |
| K. marxianus PYCC 3286 | CBS 607; ATCC 4135; CECT 1018; DBVPG 6161; JCM 3760; NCYC 6 |
| K. marxianus PYCC 3510 | NRRL Y-1122 |
| K. marxianus PYCC 3884 | CBS 397; ATCC 46537; ATCC 56497; ATCC 56752; CCRC 21477; CCRC 21698; CCT 4086; CDBB 910; CDBB 946; CECT 10584; DBVPG 6164; DSM 5422; IAM 12491; IFO 1735; JCM 22013; KCM 0230; KCTC 7152; KCTC 7155; MCYC 2712; MUCL 30017; NBIMCC 3585; NBRC 1735; NCYC 851; NCYC 1425; NRRL Y-2415; UCD 71-58; VTT C-81107; VTT C-81108 |
| Parameter | NF Permeate | RO Permeate | Mandatory or Recommended Values (Table S1 2) |
|---|---|---|---|
| pH (18 °C) | 5.85 | 5.39 | 6.5–8.5 |
| Total dissolved solids (g/L) (180 °C) | 3.86 | 0.19 | <0.5–0.6 |
| Conductivity (µS/cm) (20 °C) | 6440 | 317 | 2500 |
| Anions (mg/L) | |||
| F- | <0.17 | <0.17 | <0.5–4.0 |
| Cl- | 1890 | 78.5 | <200–250 3 |
| HCO3- | 105 | 20.7 | |
| SO42- | <4.0 | <4.0 | <250–500 3 |
| H2PO4- | 92.4 | 4.8 | |
| NO3- | 4.0 | <1.0 | <10–50 |
| NO2- | 0.021 | <0.005 | <0.5–10 |
| Cations (mg/L) | |||
| Na+ | 725 | 18.7 | <200 3 |
| K+ | 911 | 73.2 | |
| Mg2+ | 6.2 | 0.2 | <300 3 |
| Ca2+ | 32.9 | 1.0 | <300 3 |
| Fe2+ | <0.063 | <0.063 | <0.2–0.3 3 |
| NH4+ | 121 | 0.13 | <0.5–1.5 3 |
| Mn2+ | <0.031 | <0.031 | <0.05–0.1 3 |
| Vestigial Elements (µg/L) | |||
| Cu | n.d. 1 | n.d. 1 | <1000–2000 |
| Zn | n.d. 1 | n.d. 1 | <1000–4000 |
| Cd | <13 | <13 | <3–10 |
| Pb | <25 | <25 | <10–15 |
| Hg | <0.08 | <0.08 | <0.5–6 |
| Parameter | NF Retentate | RO Retentate | Reference Media (Table S2 2) |
|---|---|---|---|
| Main components (g/L) | |||
| Main Sugar | 129 ± 7 (lactose) | 145 ± 6 (lactose) | 5–22 (d-glucose) |
| Galactose | 1.7 ± 0.1 | 1.7 ± 0.2 | |
| Citric Acid | 15 ± 1 | 18 ± 2 | |
| Lactic Acid | 1.6 ± 0.3 | 0.9 ± 0.3 | |
| Proteins | 14 ± 2 | 18 ± 1 | |
| pH (25 °C) | 5.34 ± 0.01 | 5.73 ± 0.03 | 5.0–6.0 |
| Anions (mg/L) | |||
| F- | 373 | 130 | |
| Cl- | 2254 | 3213 | 0.4–125 |
| HCO3- | 3356 | 3280 | |
| SO42- | 399 | 382 | 3827–5657 |
| H2PO4- | 1938 | 2084 | 708–10262 |
| NO3- | <5 | <5 | |
| NO2- | <0.005 | <0.005 | |
| MoO42- | n.d. 1 | n.d. 1 | 0.2–0.6 |
| BO3- | n.d. 1 | n.d. 1 | 0.5–1.9 |
| Cations (mg/L) | |||
| Li+ | <0.03 | <0.03 | |
| Na+ | 2067 | 1139 | 0.1–39 |
| K+ | 1975 | 4476 | 285–4137 |
| Mg2+ | 178 | 189 | 493 |
| Ca2+ | 436 | 442 | 1.2–36 |
| Fe (total) | <0.10 | <0.10 | 0.1–1.2 |
| NH4+ | 148 | 222 | 1361–2045 |
| Mn2+ | <0.05 | <0.05 | 0.2–0.5 |
| Vestigial Elements (µg/L) | |||
| Cu | 68 | 31 | 16–127 |
| Zn | 265 | 203 | 131–2041 |
| Co | <60 | <60 | 0–118 |
| Strains | Time of Maximum Ethanol | Residual Lactose (g/L) 1 | Ethanol Titer (g/L) 1 | Acetic Acid (g/L) 2 | Glycerol (g/L) 2 | Ethanol Yield (g/g) 1 |
|---|---|---|---|---|---|---|
| Nanofiltration (NF) | ||||||
| K. marxianus PYCC 3286 | 16 h | 0 | 56 ± 0 | 2.7 ± 0.2 | 0.7 ± 0.3 | 0.50 ± 0.00 |
| K. lactis PYCC 4356 | 16 h | 0 | 52 ± 2 | 0.6 ± 0.4 | 0.8 ± 0.1 | 0.49 ± 0.01 |
| K. lactis PYCC 4357 | 20 h | 4 ± 4 (6 ± 2) | 49 ± 3 (46 ± 2) | 1.4 ± 0.4 | 1.5 ± 0.2 | 0.48 ± 0.01 (0.47 ± 0.00) |
| K. marxianus PYCC 3510 | 24 h | 16 ± 14 (29 ± 4) | 38 ± 1 (31 ± 2) | 4.2 ± 0.0 | 0.1 ± 0.1 | 0.42 ± 0.04 (0.39 ± 0.02) |
| K. lactis PYCC 3889 | 46 h | 0 (42 ± 3) | 39 ± 1 (32 ± 0) | 0.5 ± 0.1 | 2.3 ± 0.6 | 0.35 ± 0.00 (0.46 ± 0.02) |
| K. lactis var. lactis PYCC 3206 | 24 h | 20 ± 0 (38 ± 2) | 41 ± 1 (33 ± 0) | 0 | 2.8 ± 0.3 | 0.45 ± 0.02 (0.44 ± 0.00) |
| K. lactis PYCC 3207 | 24 h | 8 ± 6 (31 ± 3) | 42 ± 2 (32 ± 1) | 0.4 ± 0.4 | 0.8 ± 0.3 | 0.44 ± 0.03 (0.45 ± 0.02) |
| K. marxianus PYCC 2948 | 24 h | 27 ± 6 (49 ± 5) | 34 ± 2 (26 ± 1) | 1.7 ± 0.3 | 0.2 ± 0.2 | 0.45 ± 0.01 (0.47 ± 0.00) |
| K. marxianus PYCC 3884 (CBS 397) | 46 h | 32 ± 9 (65 ± 6) | 26 ± 2 (21 ± 2) | 8.3 ± 0.7 | 0 | 0.31 ± 0.03 (0.41 ± 0.04) |
| K. marxianus PYCC 3282 | 20 h | 94 ± 8 (95 ± 6) | 8 ± 1 (7 ± 0) | 3.1 ± 0.2 | 0 | 0.36 ± 0.02 (0.36 ± 0.02) |
| Reverse Osmosis (RO) | ||||||
| K. marxianus PYCC 3286 | 16 h | 0 | 59 ± 1 | 1.8 ± 0.3 | 1.3 ± 0.4 | 0.48 ± 0.01 |
| K. lactis PYCC 4356 | 16 h | 0 | 57 ± 3 | 0.5 ± 0.4 | 1.7 ± 0.2 | 0.47 ± 0.02 |
| K. lactis PYCC 4357 | 24 h | 13 ± 5 (24 ± 1) | 49 ± 4 (42 ± 2) | 2.2 ± 0.2 | 2.6 ± 0.4 | 0.45 ± 0.04 (0.44 ± 0.00) |
| K. marxianus PYCC 3510 | 24 h | 6 ± 6 (22 ± 4) | 50 ± 0 (41 ± 0) | 4.3 ± 0.3 | 0.8 ± 0.2 | 0.46 ± 0.01 (0.45 ± 0.02) |
| K. lactis PYCC 3889 | 46 h | 0 (42 ± 1) | 39 ± 2 (34 ± 2) | 0.3 ± 0.3 | 2.7 ± 0.1 | 0.32 ± 0.01 (0.42 ± 0.00) |
| K. lactis var. lactis PYCC 3206 | 24 h | 33 ± 5 (50 ± 2) | 42 ± 1 (34 ± 0) | 0 | 3.0 ± 1.0 | 0.45 ± 0.02 (0.45 ± 0.00) |
| K. lactis PYCC 3207 | 24 h | 18 ± 12 (43 ± 7) | 47 ± 0 (33 ± 1) | 0.3 ± 0.3 | 1.0 ± 0.7 | 0.46 ± 0.03 (0.42 ± 0.02) |
| K. marxianus PYCC 2948 | 24 h | 20 ±8 (51 ± 9) | 44 ± 3 (28 ± 1) | 1.5 ± 0.2 | 0.8 ± 0.0 | 0.45 ± 0.01 (0.42 ± 0.02) |
| K. marxianus PYCC 3884 (CBS 397) | 24 h | 11 ± 10 (32 ± 5) | 48 ± 3 (41 ± 2) | 4.3 ± 0.5 | 0.5 ± 0.2 | 0.42 ± 0.02 (0.44 ± 0.01) |
| K. marxianus PYCC 3282 | 20 h | 111 ± 6 (112 ± 4) | 8 ± 2 (8 ± 2) | 2.2 ± 0.3 | 0.1 ± 0.1 | 0.39 ± 0.02 (0.40 ± 0.01) |
| Strains | Oxygen- Limiting Condition | Time of Maximum Ethanol 1 | Consumed Lactose (%) 1 | Ethanol Titer (g/L) 1 | Maximum Ethanol Yield (g/g) | Ethanol Productivity (g/L/h) 2 | Lactose Consumption Rate (g/L/h) 2 |
|---|---|---|---|---|---|---|---|
| Nanofiltration (NF) | |||||||
| K. marxianus PYCC 3286 | Mild | 16 h | 98 ± 2 | 52 ± 4 | 0.47 ± 0.04 | 3.51 ± 0.04 | 8.24 ± 0.53 |
| Severe | 30 h | 100 ± 0 | 61 ± 1 | 0.49 ± 0.02 | 3.26 ± 0.28 | 7.01 ± 0.78 | |
| K. lactis PYCC 4356 | Mild | 16 h | 98 ± 2 | 48 ± 1 | 0.46 ± 0.02 | 3.93 ± 0.05 | 9.01 ± 0.60 |
| Severe | 55 h (120 h) | 88 ± 1 (99 ± 1) | 49 ± 0 (58 ± 1) | 0.47 ± 0.02 | 3.19 ± 0.14 | 6.87 ± 0.30 | |
| K. marxianus PYCC 3884 (CBS 397) | Mild | 48 h | 78 ± 3 | 26 ± 1 | 0.28 ± 0.02 | 1.52 ± 0.14 | 3.73 ± 0.09 |
| Severe | 55 h (120 h) | 72 ± 2 (100 ± 0) | 41 ± 1 (62 ± 0) | 0.46 ± 0.02 | 1.32 ± 0.12 | 3.19 ± 0.20 | |
| Reverse Osmosis (RO) | |||||||
| K. marxianus PYCC 3286 | Mild | 16 h | 99 ± 1 | 58 ± 1 | 0.46 ± 0.02 | 4.18 ± 0.01 | 9.57 ± 0.63 |
| Severe | 29 h | 100 ± 0 | 70 ± 2 | 0.49 ± 0.01 | 3.67 ± 0.31 | 8.78 ± 0.41 | |
| K. lactis PYCC 4356 | Mild | 20 h | 98 ± 2 | 52 ± 1 | 0.44 ± 0.01 | 4.08 ± 0.04 | 9.67 ± 0.60 |
| Severe | 54 h (120 h) | 94 ± 1 (100 ± 0) | 60 ± 1 (67 ± 2) | 0.47 ± 0.01 | 3.75 ± 0.25 | 8.30 ± 0.46 | |
| K. marxianus PYCC 3884 (CBS 397) | Mild | 30 h | 89 ± 2 | 39 ± 5 | 0.32 ± 0.04 | 2.74 ± 0.11 | 6.69 ± 0.54 |
| Severe | 54 h (120 h) | 90 ± 4 (100 ± 0) | 59 ± 6 (70 ± 2) | 0.48 ± 0.03 | 2.03 ± 0.13 | 4.90 ± 0.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leandro, M.J.; Marques, S.; Ribeiro, B.; Santos, H.; Fonseca, C. Integrated Process for Bioenergy Production and Water Recycling in the Dairy Industry: Selection of Kluyveromyces Strains for Direct Conversion of Concentrated Lactose-Rich Streams into Bioethanol. Microorganisms 2019, 7, 545. https://doi.org/10.3390/microorganisms7110545
Leandro MJ, Marques S, Ribeiro B, Santos H, Fonseca C. Integrated Process for Bioenergy Production and Water Recycling in the Dairy Industry: Selection of Kluyveromyces Strains for Direct Conversion of Concentrated Lactose-Rich Streams into Bioethanol. Microorganisms. 2019; 7(11):545. https://doi.org/10.3390/microorganisms7110545
Chicago/Turabian StyleLeandro, Maria José, Susana Marques, Belina Ribeiro, Helena Santos, and César Fonseca. 2019. "Integrated Process for Bioenergy Production and Water Recycling in the Dairy Industry: Selection of Kluyveromyces Strains for Direct Conversion of Concentrated Lactose-Rich Streams into Bioethanol" Microorganisms 7, no. 11: 545. https://doi.org/10.3390/microorganisms7110545
APA StyleLeandro, M. J., Marques, S., Ribeiro, B., Santos, H., & Fonseca, C. (2019). Integrated Process for Bioenergy Production and Water Recycling in the Dairy Industry: Selection of Kluyveromyces Strains for Direct Conversion of Concentrated Lactose-Rich Streams into Bioethanol. Microorganisms, 7(11), 545. https://doi.org/10.3390/microorganisms7110545






