Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro
Abstract
1. Introduction
2. Methods
2.1. Cell Lines, Cell Culture, and Transfection
2.2. CRISPR/Cas9-Mediated Targeting of APOBEC3s
2.3. Isolation of Nucleic Acids, Reverse Transcription, and PCR Analysis
2.4. Southern Blot Analysis
2.5. HBsAg Quantification
2.6. Immunocytochemistry and Fluorescent Microscopy
2.7. FACS Analysis
2.8. Statistics
3. Results
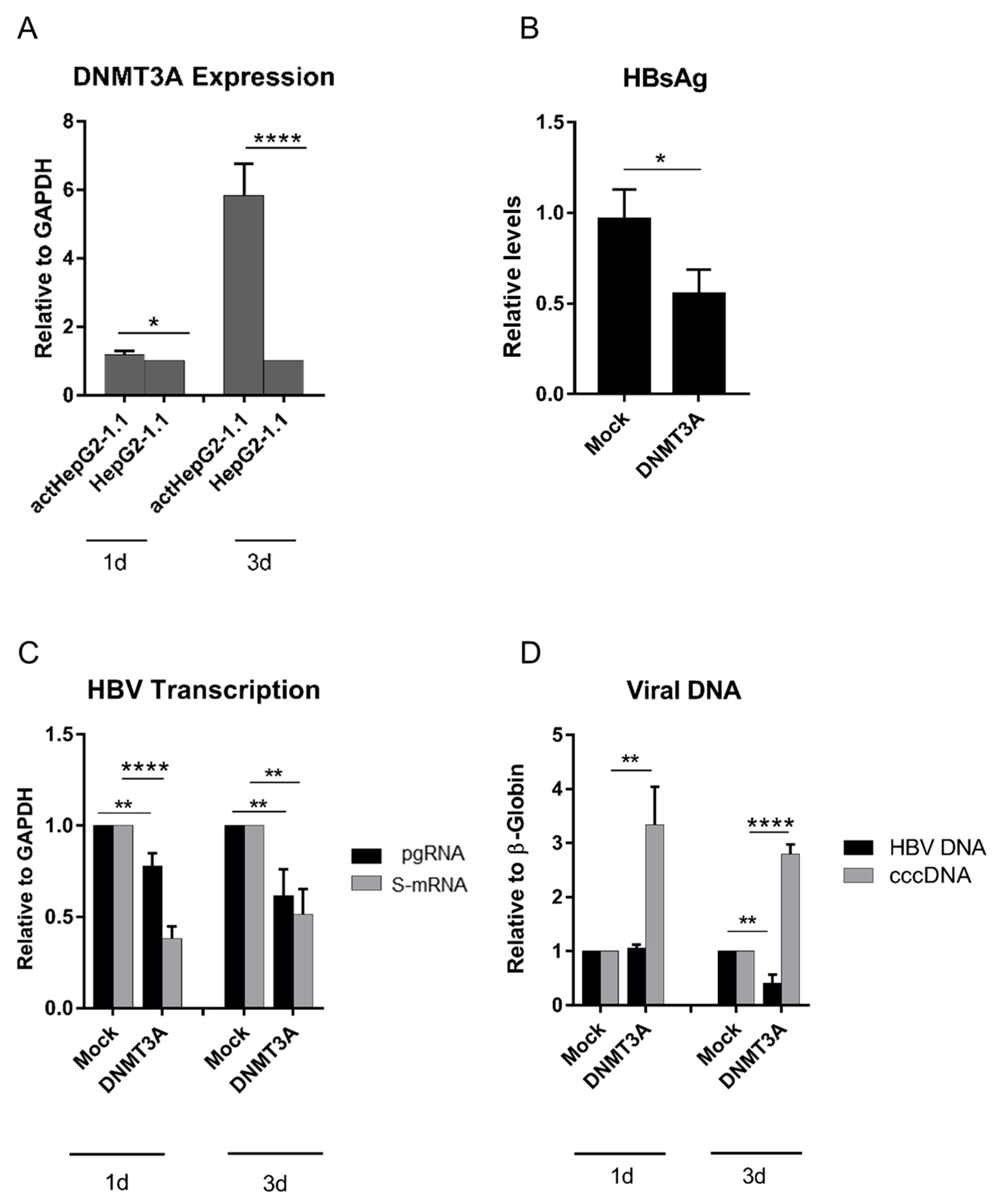
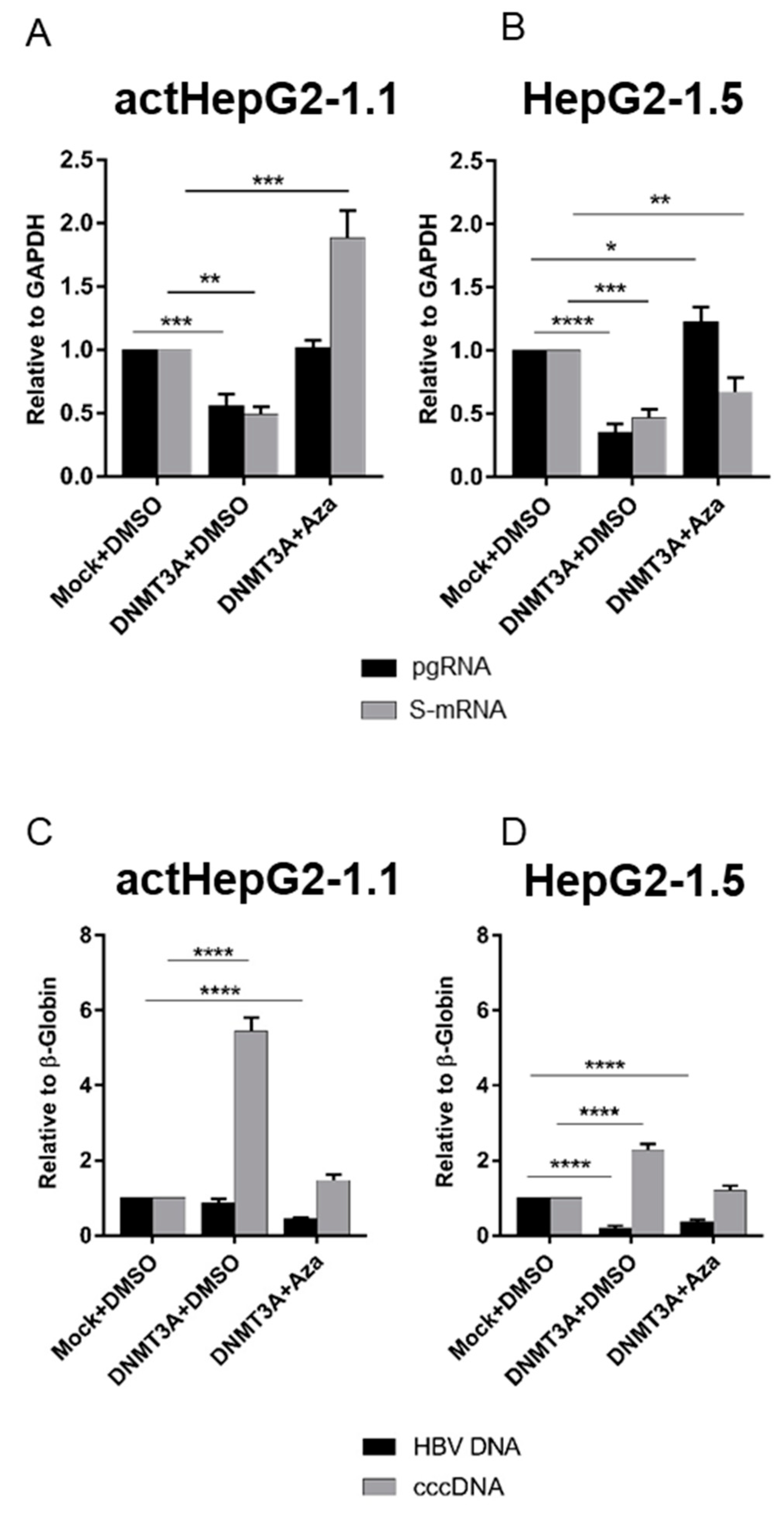
3.1. Overexpression of DNMT3A Suppresses HBV Replication But Increases cccDNA Levels
3.2. DNMT3A Stimulates Cell Cycling in HBV-Infected Cells
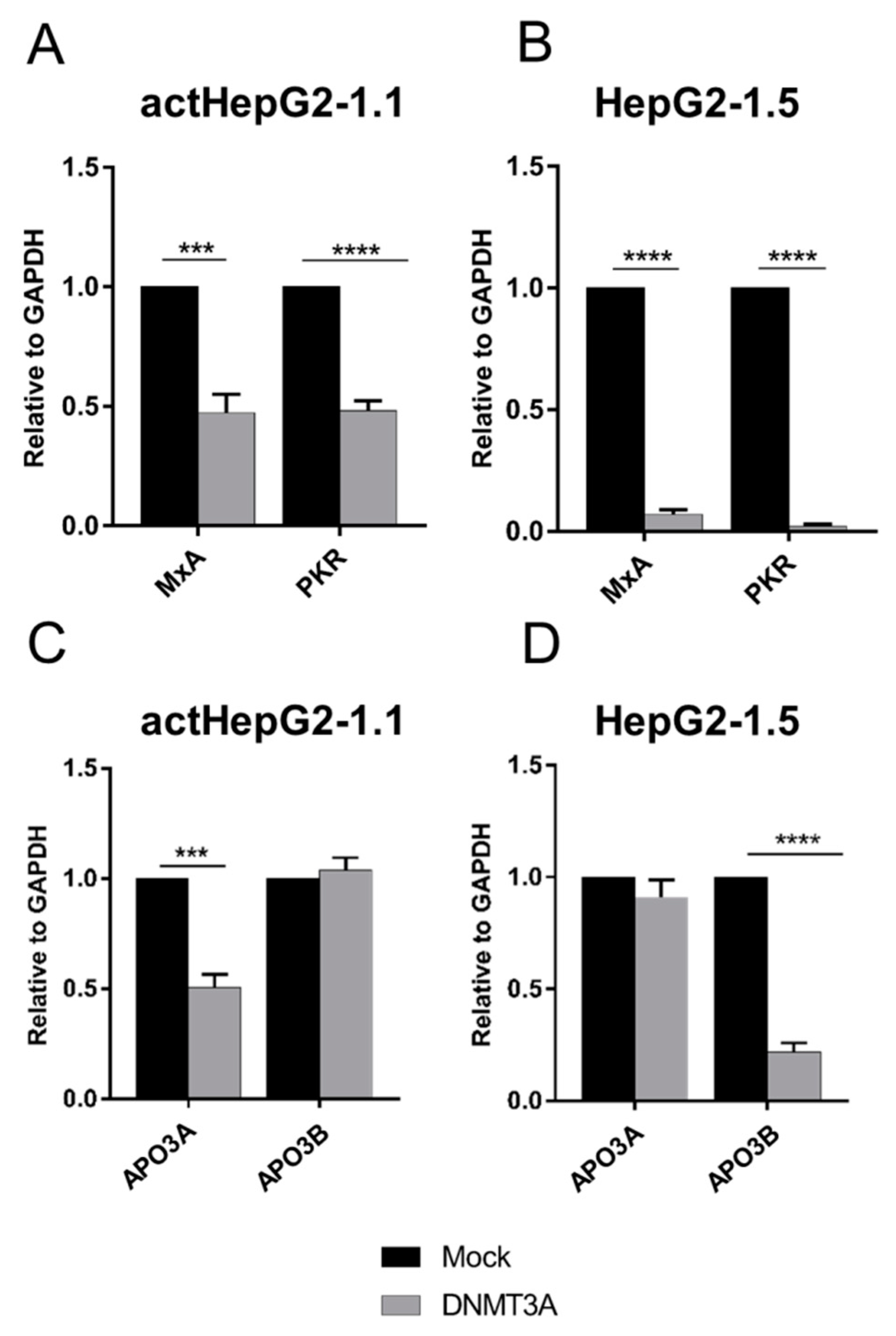
3.3. DNMT3A Downregulates Expression of HBV Restriction Factors Including APOBEC3s
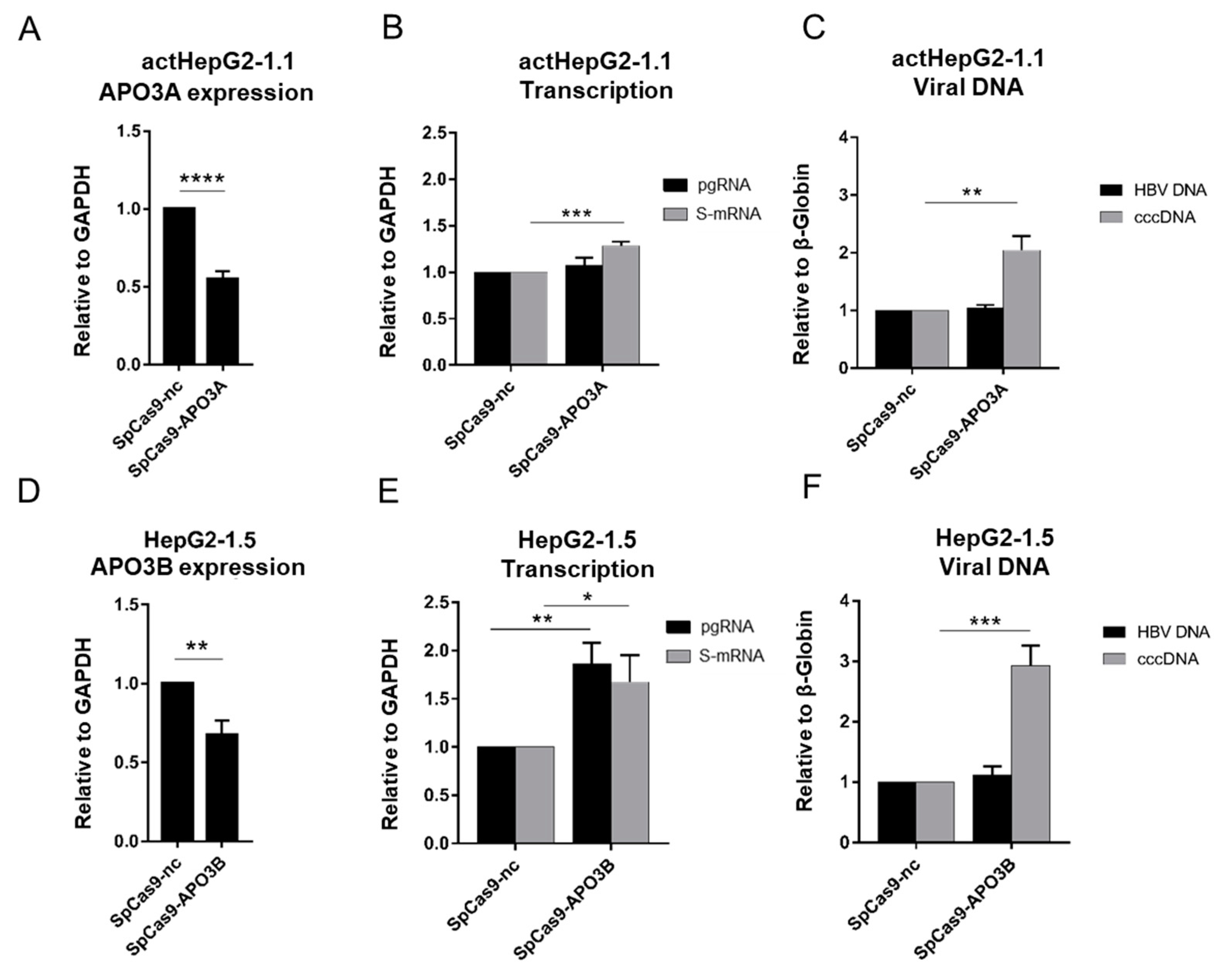
3.4. CRISPR/Cas9-Mediated Silencing of APO3A or APO3B Increases cccDNA Pool
3.5. Overexpressed DNMT3A Reduces Expression of DNA-Damage Response Factors and Aggravates HBV-Related Genome Damage
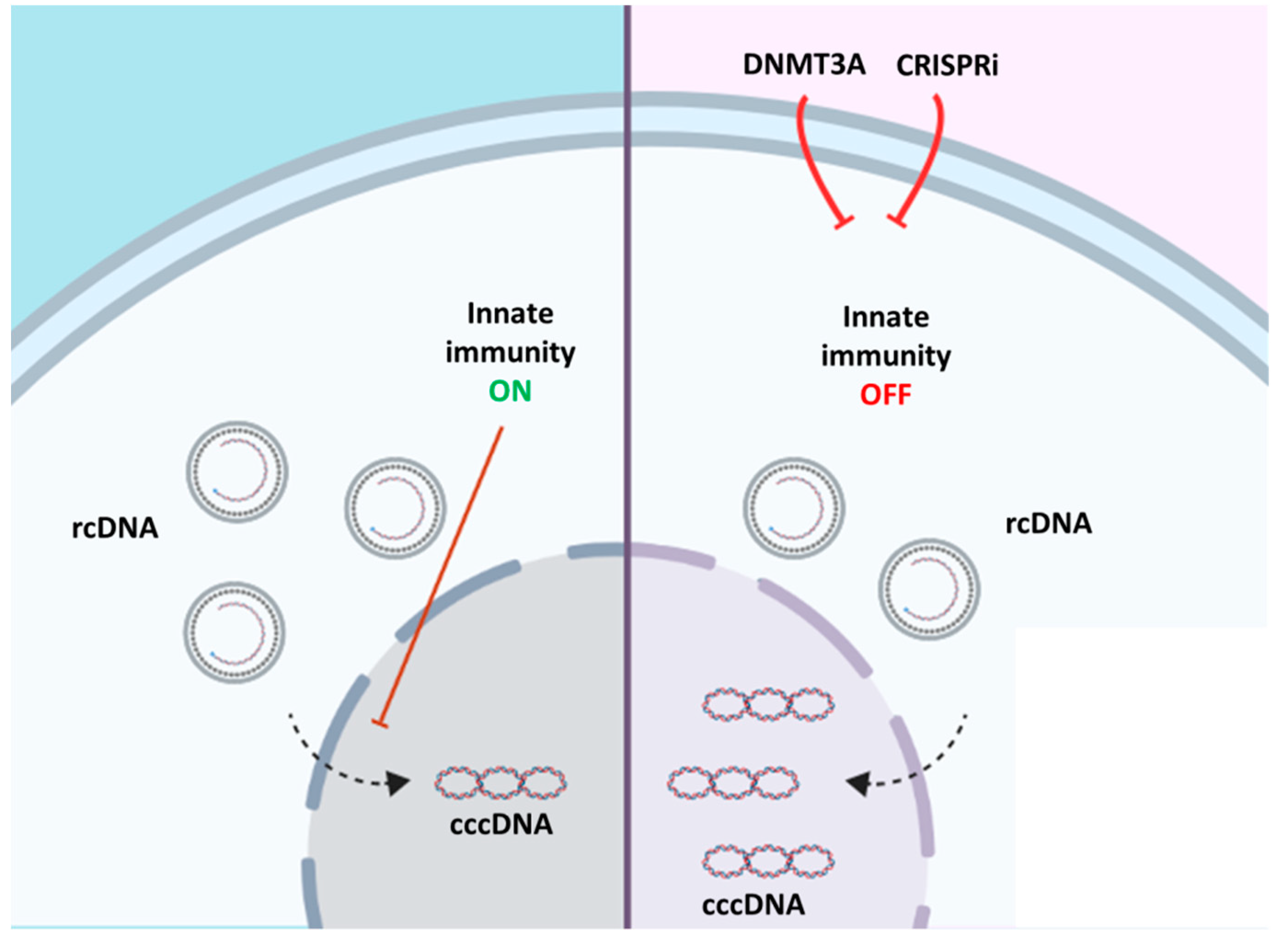
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ganem, D.; Prince, A.M. Hepatitis B virus infection—Natural history and clinical consequences. N. Engl. J. Med. 2004, 350, 1118–1129. [Google Scholar] [CrossRef]
- Nassal, M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015, 64, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-C.; Kao, J.-H. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg. Microbes Infect. 2014, 3, e64. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Nassal, M. A Role for the Host DNA Damage Response in Hepatitis B Virus cccDNA Formation—And Beyond? Viruses 2017, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Lutgehetmann, M.; Volz, T.; Koepke, A.; Broja, T.; Tigges, E.; Lohse, A.W.; Fuchs, E.; Murray, J.M.; Petersen, J.; Dandri, M. In Vivo Proliferation of Hepadnavirus-Infected Hepatocytes Induces Loss of Covalently Closed Circular DNA in Mice. Hepatology 2010, 52, 16–24. [Google Scholar] [CrossRef]
- Allweiss, L.; Volz, T.; Giersch, K.; Kah, J.; Raffa, G.; Petersen, J.; Lohse, A.W.; Beninati, C.; Pollicino, T.; Urban, S.; et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 2018, 67, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Chakraborty, A.; Chou, W.-M.; Hasreiter, J.; Wettengel, J.M.; Stadler, D.; Bester, R.; Asen, T.; Zhang, K.; Wisskirchen, K. Hepatitis B virus (HBV) genome recycling and de novo secondary infection events maintain stable cccDNA levels. J. Hepatol. 2018, 69, 1231–1241. [Google Scholar] [CrossRef]
- Busca, A.; Kumar, A. Innate immune responses in hepatitis B virus (HBV) infection. Virol. J. 2014, 11, 22. [Google Scholar] [CrossRef]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Nair, S.; Zlotnick, A. Asymmetric Modification of Hepatitis B Virus (HBV) Genomes by an Endogenous Cytidine Deaminase inside HBV Cores Informs a Model of Reverse Transcription. J. Virol. 2018, 92, e02190-17. [Google Scholar] [CrossRef]
- Vivekanandan, P.; Thomas, D.; Torbenson, M. Methylation regulates hepatitis B viral protein expression. J. Infect. Dis. 2009, 199, 1286–1291. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, Y.; Mu, S.; Zhang, J.; Yan, Z. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J. Med. Virol. 2009, 81, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mao, R.; Yan, R.; Cai, D.; Zhang, Y.; Zhu, H.; Kang, Y.; Liu, H.; Wang, J.; Qin, Y.; et al. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS ONE 2014, 9, e110442. [Google Scholar] [CrossRef] [PubMed]
- Kostyushev, D.S.; Zueva, A.P.; Brezgin, S.A.; Lipatnikov, A.D.; Simirskii, V.N.; Glebe, D.; Volchkova, E.V.; Shipulin, G.A.; Chulanov, V.P. Overexpression of DNA-methyltransferases in persistency of cccDNA pool in chronic hepatitis B. Ter. Arkhiv 2017, 89, 21–26. [Google Scholar] [CrossRef]
- Vivekanandan, P.; Daniel, H.D.-J.; Kannangai, R.; Martinez-Murillo, F.; Torbenson, M. Hepatitis B virus replication induces methylation of both host and viral DNA. J. Virol. 2010, 84, 4321–4329. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar]
- Kim, S.; Lee, H.-S.; Ji, J.-H.; Cho, M.-Y.; Yoo, Y.-S.; Park, Y.-Y.; Cha, H.-J.; Lee, Y.; Kim, Y.; Cho, H. Hepatitis B virus X protein activates the ATM-Chk2 pathway and delays cell cycle progression. J. Gen. Virol. 2015, 96, 2242–2251. [Google Scholar] [CrossRef]
- Becker, S.A.; Lee, T.-H.; Butel, J.S.; Slagle, B.L. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 1998, 72, 266–272. [Google Scholar]
- Hsieh, Y.-H.; Su, I.-J.; Wang, H.-C.; Chang, W.-W.; Lei, H.-Y.; Lai, M.-D.; Chang, W.-T.; Huang, W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis 2004, 25, 2023–2032. [Google Scholar] [CrossRef]
- Tarocchi, M.; Polvani, S.; Marroncini, G.; Galli, A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World J. Gastroenterol. 2014, 20, 11630. [Google Scholar] [CrossRef]
- Kostyushev, D.; Brezgin, S.; Kostyusheva, A.; Zarifyan, D.; Goptar, I.; Chulanov, V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell. Mol. Life Sci. 2019, 76, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Kostyushev, D.; Kostyusheva, A.; Brezgin, S.; Zarifyan, D.; Utkina, A.; Goptar, I.; Chulanov, V. Suppressing the NHEJ pathway by DNA-PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci. Rep. 2019, 9, 1847. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.P.; Kostyushev, D.S.; Brezgin, S.A.; Zarifyan, D.N.; Volchkova, E.V.; Chulanov, V.P. Small Molecular Inhibitors of DNA Double Strand Break Repair Pathways Increase the ANTI-HBV Activity of CRISPR/Cas9. Mol. Biol. 2019, 53, 274–285. [Google Scholar] [CrossRef]
- Stemmer, M.; Thumberger, T.; del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-T.; Han, T.; Li, Y.; Yang, B.; Wang, Y.-J.; Wang, F.; Jing, X.; Du, Z. Enhanced specificity of real-time PCR for measurement of hepatitis B virus cccDNA using restriction endonuclease and plasmid-safe ATP-dependent DNase and selective primers. J. Virol. Methods 2010, 169, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Nie, H.; Yan, R.; Guo, J.-T.; Block, T.M.; Guo, H. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol. Biol. 2013, 1030, 151–161. [Google Scholar] [PubMed]
- Kostyushev, D.S.; Brezgin, S.A.; Kostyusheva, A.P.; Lipatnikov, A.D.; Simirskii, V.N.; Mamonova, N.A.; Volchkova, E.V.; Maleyev, V.V.; Chulanov, V. Increased formation of phosphorylated H2AX foci in nuclei of cells infected by hepatitis B AND B+D viruses. Vopr. Virusol. 2018, 63, 165–170. [Google Scholar]
- Chong, C.-L.; Chen, M.-L.; Wu, Y.-C.; Tsai, K.-N.; Huang, C.-C.; Hu, C.-P.; Jeng, K.-S.; Chou, Y.-C.; Chang, C. Dynamics of HBV cccDNA expression and transcription in different cell growth phase. J. Biomed. Sci. 2011, 18, 96. [Google Scholar] [CrossRef]
- Guo, H.; Mao, R.; Block, T.M.; Guo, J.-T. Production and Function of the Cytoplasmic Deproteinized Relaxed Circular DNA of Hepadnaviruses. J. Virol. 2010, 84, 387–396. [Google Scholar] [CrossRef]
- Yeh, C.T.; Chiu, H.T.; Chu, C.M.; Liaw, Y.F. G(1) phase dependent nuclear localization of relaxed-circular hepatitis B virus DNA and aphidicolin-induced accumulation of covalently closed circular DNA. J. Med. Virol. 1998, 55, 42–50. [Google Scholar] [CrossRef]
- Sirma, H.; Giannini, C.; Poussin, K.; Paterlini, P.; Kremsdorf, D.; Brechot, C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 1999, 18, 4848–4859. [Google Scholar] [CrossRef] [PubMed]
- Gearhart, T.L.; Bouchard, M.J. Replication of the hepatitis B virus requires a calcium-dependent HBx-induced G1 phase arrest of hepatocytes. Virology 2010, 407, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Han, X.; Guan, G.; Wu, N.; Sun, J.; Pak, V.; Liang, G. TGF-beta triggers HBV cccDNA degradation through AID-dependent deamination. FEBS Lett. 2016, 590, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.-L.; Zhang, L.; Cheng, N.; Xu, X.; Deng, Q.; Teng, X.-M.; Wang, K.-S.; Zhang, X.; Huang, J.; Han, Z.-G. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J. Hepatol. 2009, 50, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Stadler, D.; Lucifora, J.; Reisinger, F.; Webb, D.; Hoesel, M.; Michler, T.; Wisskirchen, K.; Cheng, X.; Zhang, K.; et al. Interferon-gamma and Tumor Necrosis Factor-alpha Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis. Gastroenterology 2016, 150, 194–205. [Google Scholar] [CrossRef]
- Gordien, E.; Rosmorduc, O.; Peltekian, C.; Garreau, F.; Bréchot, C.; Kremsdorf, D. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 2001, 75, 2684–2691. [Google Scholar] [CrossRef]
- Park, I.-H.; Baek, K.-W.; Cho, E.-Y.; Ahn, B.-Y. PKR-dependent mechanisms of interferon-α for inhibiting hepatitis B virus replication. Mol. Cells 2011, 32, 167–172. [Google Scholar] [CrossRef]
- Oh, B.-K.; Kim, H.; Park, H.-J.; Shim, Y.-H.; Choi, J.; Park, C.; Park, Y.N. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int. J. Mol. Med. 2007, 20, 65–73. [Google Scholar] [CrossRef]
- Ren, J.-H.; Chen, X.; Zhou, L.; Tao, N.-N.; Zhou, H.-Z.; Liu, B.; Li, W.-Y.; Huang, A.-L.; Chen, J. Protective Role of Sirtuin3 (SIRT3) in Oxidative Stress Mediated by Hepatitis B Virus X Protein Expression. PLoS ONE 2016, 11, e0150961. [Google Scholar] [CrossRef]
- Matsuda, Y.; Wakai, T.; Kubota, M.; Osawa, M.; Takamura, M.; Yamagiwa, S.; Aoyagi, Y.; Sanpei, A.; Fujimaki, S. DNA Damage Sensor γ-H2AX Is Increased in Preneoplastic Lesions of Hepatocellular Carcinoma. Sci. World J. 2013, 2013, 597095. [Google Scholar] [CrossRef]
- Teodoridis, J.M.; Strathdee, G.; Brown, R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist. Updates 2004, 7, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chu, Y.; Ma, H.; Zhang, Y.; Zhang, X.; Zhao, D.; Li, Z.; Wang, J.; Gao, Y.; Xiao, L.; et al. Epigenetic Interventions Increase the Radiation Sensitivity of Cancer Cells. Curr. Pharm. Des. 2014, 20, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Hou, N.-B.; Song, T.; He, X.; Zheng, Z.-R.; Ma, Q.-J.; Li, L.; Zhang, Y.-H.; Zhong, H. Cellular DNA repair cofactors affecting hepatitis B virus infection and replication. World J. Gastroenterol. 2008, 14, 5059–5065. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Brezgin, S.; Bayurova, E.; Gordeychuk, I.; Isaguliants, M.; Goptar, I.; Urusov, F.; Nikiforova, A.; Volchkova, E.; Kostyushev, D.; et al. ATM and ATR Expression Potentiates HBV Replication and Contributes to Reactivation of HBV Infection upon DNA Damage. Viruses 2019, 11, 997. [Google Scholar] [CrossRef]
- Matsuda, Y.; Ichida, T. Impact of hepatitis B virus X protein on the DNA damage response during hepatocarcinogenesis. Med. Mol. Morphol. 2009, 42, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Sohn, B.H.; Yu, E.; Suh, D.J.; Chung, Y.H.; Lee, J.H.; Surzycki, S.J.; Lee, Y.I. Aberrant Epigenetic Modifications in Hepatocarcinogenesis Induced by Hepatitis B Virus X Protein. Gastroenterology 2007, 132, 1476–1494. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandan, P.; Thomas, D.; Torbenson, M. Hepatitis B viral DNA is methylated in liver tissues. J. Viral Hepat. 2008, 15, 103–107. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483. [Google Scholar] [CrossRef]
- Cheng, X.; Xia, Y.; Serti, E.; Block, P.D.; Chung, M.; Chayama, K.; Rehermann, B.; Liang, T.J. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology 2017, 66, 1779–1793. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Zhang, X.-Y. Innate immune recognition of hepatitis B virus. World J. Hepatol. 2015, 7, 2319–2322. [Google Scholar] [CrossRef]
- Mutz, P.; Metz, P.; Lempp, F.A.; Bender, S.; Qu, B.; Schöneweis, K.; Seitz, S.; Tu, T.; Restuccia, A.; Frankish, J. HBV bypasses the innate immune response and does not protect HCV from antiviral activity of interferon. Gastroenterology 2018, 154, 1791–1804. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Sohn, J.A. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol. Ther. 2016, 24, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sohn, J.A.; Seeger, C. Distribution of Hepatitis B Virus Nuclear DNA. J. Virol. 2018, 92, e01391-17. [Google Scholar] [CrossRef] [PubMed]
- Perlman, D.H.; Berg, E.A.; O’connor, P.B.; Costello, C.E.; Hu, J. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. USA 2005, 102, 9020–9025. [Google Scholar] [CrossRef] [PubMed]
- Kuss-Duerkop, S.K.; Westrich, J.A.; Pyeon, D. DNA Tumor Virus Regulation of Host DNA Methylation and Its Implications for Immune Evasion and Oncogenesis. Viruses 2018, 10, 82. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brezgin, S.; Kostyusheva, A.; Bayurova, E.; Gordeychuk, I.; Isaguliants, M.; Goptar, I.; Nikiforova, A.; Smirnov, V.; Volchkova, E.; Glebe, D.; et al. Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro. Microorganisms 2019, 7, 533. https://doi.org/10.3390/microorganisms7110533
Brezgin S, Kostyusheva A, Bayurova E, Gordeychuk I, Isaguliants M, Goptar I, Nikiforova A, Smirnov V, Volchkova E, Glebe D, et al. Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro. Microorganisms. 2019; 7(11):533. https://doi.org/10.3390/microorganisms7110533
Chicago/Turabian StyleBrezgin, Sergey, Anastasiia Kostyusheva, Ekaterina Bayurova, Ilya Gordeychuk, Maria Isaguliants, Irina Goptar, Anastasiia Nikiforova, Valery Smirnov, Elena Volchkova, Dieter Glebe, and et al. 2019. "Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro" Microorganisms 7, no. 11: 533. https://doi.org/10.3390/microorganisms7110533
APA StyleBrezgin, S., Kostyusheva, A., Bayurova, E., Gordeychuk, I., Isaguliants, M., Goptar, I., Nikiforova, A., Smirnov, V., Volchkova, E., Glebe, D., Kostyushev, D., & Chulanov, V. (2019). Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro. Microorganisms, 7(11), 533. https://doi.org/10.3390/microorganisms7110533






