Aided Phytoremediation to Clean Up Dioxins/Furans-Aged Contaminated Soil: Correlation between Microbial Communities and Pollutant Dissipation
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Setup
2.2. Arbuscular Mycorrhizal Root Colonization
2.3. Determination of Plant Growth
2.4. Estimation of Pigment Contents
2.5. Extraction, Estimation of Soil PCDD/F Toxic Equivalent Values
2.6. Soil Enzyme Assays
2.6.1. Soil Dehydrogenase Activity
2.6.2. Soil Fluorescein Diacetate Hydrolase
2.7. Cytotoxicity Tests
2.8. Soil DNA Extraction and DNA Quantification
2.9. PCR and Amplicon Sequencing
2.10. Bioinformatic Analysis
2.11. Statistical Analysis
3. Results
3.1. Mycorrhizal Root Colonization and Root Growth of Alfalfa
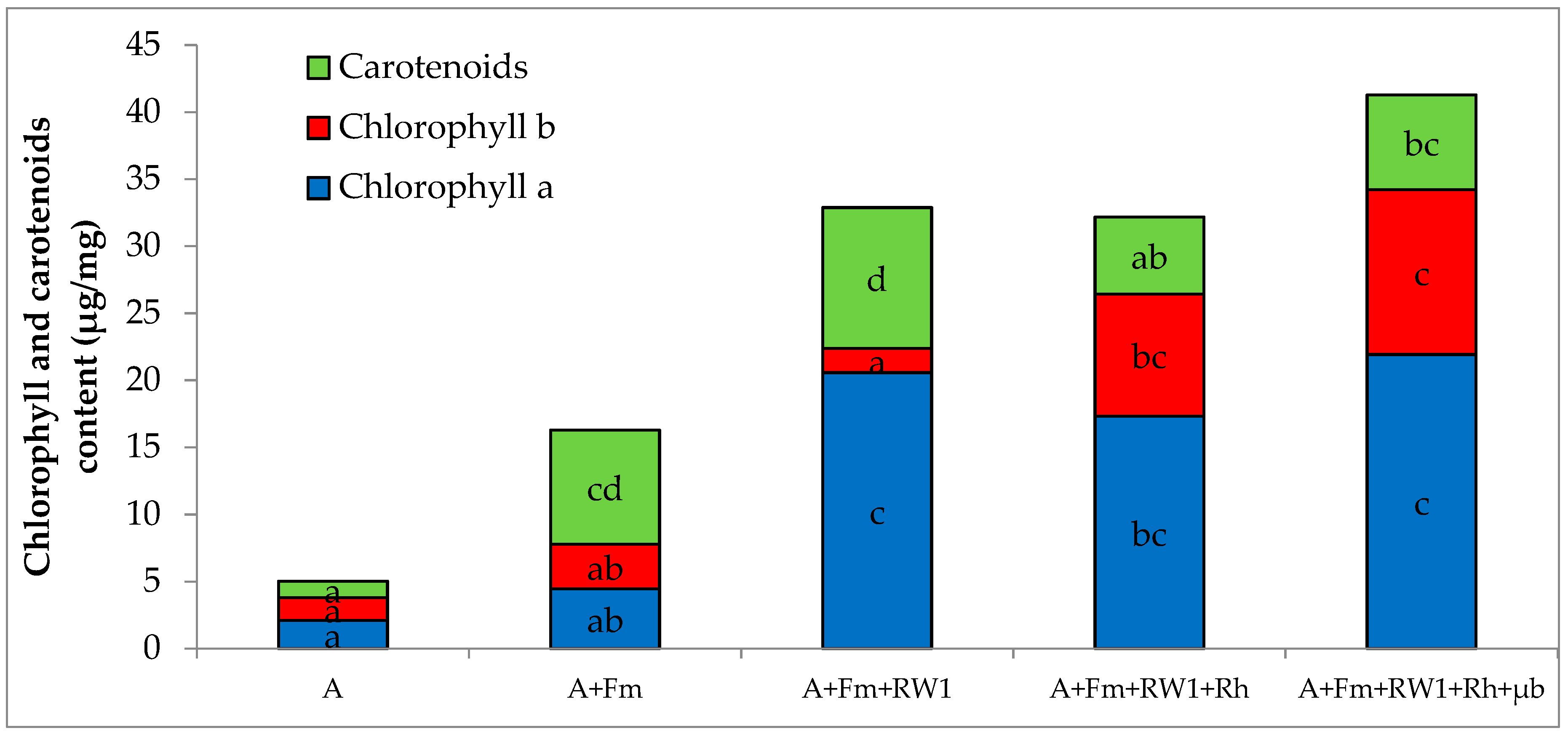
3.2. Shoot Growth and Photosynthetic Pigment Content of Alfalfa
3.3. Total PCDD/F Dissipation
3.4. Estimation of the Polluted Soil Cytotoxicity after phytoremediation with A+Fm+RW1+Rh+µb Condition
3.5. Assessment of Soil Microbial Activity
3.6. Soil Microbial Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Häggblom, M.M.; Bossert, I.D. Halogenated organic compounds: A global perspective. In Dehalogenation: Microbial Processes and Environmental Applications; Häggblom, M.M., Bossert, I.D., Eds.; Kluwer Academic: New York, NY, USA, 2003; pp. 3–29. [Google Scholar]
- Rocha, D.A.M.; Torres, J.P.M.; Reichel, K.; Novotny, E.H.; Estrella, L.F.; Medeiros, R.O.; Pereira Netto, A.D. Determination of polychlorinated dibenzo-p-dioxins and dibenzofurans PCDD/Fs in Brazilian cow milk. Sci. Total Environ. 2016, 572, 177–184. [Google Scholar] [CrossRef]
- Hanano, A.; Almousally, I.; Shaban, M.; Moursel, N.; Shahadeh, A.; Alhajji, E. Differential tissue accumulation of 2,3,7,8-Tetrachlorinated dibenzo-p-dioxin in Arabidopsis thaliana affects plant chronology, lipid metabolism and seed yield. BMC Plant Biol. 2015, 15, 193. [Google Scholar] [CrossRef]
- Budzinski, H.; Fraisse, D.; Hoogenboom, L.A.P. Dioxins in the Environment: What Are the Health Risks? INSERM Collective Expertise Centre: Paris, France, 2000; p. 1.
- Knutsen, H.K.; Alexander, J.; Barregard, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Edler, L.; Grasl-Kraupp, B. Risk for animal and human health related to the presence of dioxins and dioxin-like PCBs in feed and food. EFSA J. 2019, 16, 5333. [Google Scholar] [CrossRef]
- Pollitt, F. Polychlorinated dibenzodioxins and polychlorinated dibenzofurans. Regul. Toxicol. Pharm. 1999, 30, 63–68. [Google Scholar] [CrossRef]
- Mench, M.; Lepp, N.; Bert, V.; Schwitzguébel, J.P.; Gawronski, S.W.; Schröder, P.; Vangronsveld, J. Successes and limitations of phytotechnologies at field scale: Outcomes, assessment and outlook from COST Action 859. J. Soils Sediments 2010, 10, 1039–1070. [Google Scholar] [CrossRef]
- Wang, Y.; Oyaizu, H. Evaluation of the phytoremediation potential of four plant species for dibenzofuran-contaminated soil. J. Hazard. Mater. 2009, 168, 760–764. [Google Scholar] [CrossRef]
- Li, Y.; Liang, F.; Zhu, Y.; Wang, F. Phytoremediation of a PCB-contaminated soil by alfalfa and tall fescue single and mixed plants cultivation. J. Soils Sediments 2013, 23, 925–931. [Google Scholar] [CrossRef]
- Xu, L.; Teng, Y.; Li, Z.G.; Norton, J.M.; Luo, Y.M. Enhanced removal of polychlorinated biphenyls from alfalfa rhizosphere soil in a field study: the impact of a rhizobial inoculum. Sci. Total Environ. 2010, 408, 1007–1013. [Google Scholar] [CrossRef]
- Teng, Y.; Sun, X.; Zhu, L.; Christie, P.; Luo, Y. Polychlorinated biphenyls in alfalfa: Accumulation, sorption and speciation in different plant parts. Int. J. Phytoremediation 2017, 19, 732–738. [Google Scholar] [CrossRef]
- Ionescu, M.; Beranova, K.; Dudkova, V.; Kochankova, L.; Demnerova, K.; Macek, T.; Mackova, M. Isolation and characterization of different plant associated bacteria and their potential to degrade polychlorinated biphenyls. Int. Biodeterior. Biodegrad. 2009, 63, 667–672. [Google Scholar] [CrossRef]
- Uhlik, O.; Musilova, L.; Ridl, J.; Hroudova, M.; Vlcek, C.; Koubek, J.; Holeckova, M.; Mackova, M.; Macek, T. Plant secondary metabolite-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl. Microbiol. Biotechnol. 2013, 97, 9245–9256. [Google Scholar] [CrossRef]
- Hanano, A.; Almousally, I.; Shaban, M. Phytotoxicity effects and biological responses of Arabidopsis thaliana to 2,3,7,8-tetrachlorinated dibenzo-p-dioxin exposure. Chemosphere 2014, 104, 76–84. [Google Scholar] [CrossRef]
- Urbaniak, M.; Wyrwicka, A.; Zieliński, M.; Mankiewicz-Boczek, J. Potential for phytoremediation of PCDD/PCDF-contaminated sludge and sediments using Cucurbitaceae plants: A pilot study. Bull. Environ. Contam. Toxicol. 2016, 97, 401–406. [Google Scholar] [CrossRef]
- Shin, K.H.; Kim, K.W.; Ahn, Y. Use of biosurfactant to remediate phenanthrene-contaminated soil by the combined solubilization-biodegradation process. J. Hazard. Mater. 2006, 137, 1831–1837. [Google Scholar] [CrossRef]
- Nayak, A.S.; Vijaykumar, M.H.; Karegoudar, T.B. Characterization of biosurfactant produced by Pseudoxanthomonas sp. PNK-04 and its application in bioremediation. Int. Biodeterior. Biodegrad. 2009, 63, 73–79. [Google Scholar] [CrossRef]
- Bharali, P.; Das, S.; Konwar, B.K.; Thakur, A.J. Crude biosurfactant from thermophilic Alcaligenes faecalis: Feasibility in petro-spill bioremediation. Int. Biodeterior. Biodegrad. 2011, 65, 682–690. [Google Scholar] [CrossRef]
- Mulligan, C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005, 133, 183–198. [Google Scholar] [CrossRef]
- Viisimaa, M.; Karpenko, O.; Novikov, V.; Trapido, M.; Goi, A. Influence of biosurfactant on combined chemical-biological treatment of PCB-contaminated soil. Chem. Eng. J. 2013, 220, 352–359. [Google Scholar] [CrossRef]
- Lenoir, I.; Fontaine, J.; Lounès-Hadj Sahraoui, A. Arbuscular mycorrhizal fungal responses to abiotic stresses: A review. Phytochemistry 2016, 123, 4–15. [Google Scholar] [CrossRef]
- Lenoir, I.; Lounès-Hadj Sahraoui, A.; Laruelle, F.; Dapé, Y.; Fontaine, J. Arbuscular mycorrhizal wheat inoculation promotes alkane and polycyclic aromatic hydrocarbon biodegradation: Microcosm experiment on aged-contaminated soil. Environ. Pollut. 2016, 213, 549–560. [Google Scholar]
- Teng, Y.; Luo, Y.; Sun, X.; Tu, C.; Xu, L.; Liu, W.; Li, Z.; Christie, P. Influence of arbuscular mycorrhiza and Rhizobium on phytoremediation by alfalfa of an agricultural soil contaminated with weathered PCBs: A field study. Int. J. Phytoremediat. 2010, 12, 516–533. [Google Scholar] [CrossRef]
- Yu, X.Z.; Wu, S.C.; Wu, F.Y.; Wong, M.H. Enhanced dissipation of PAHs from soil using mycorrhizal ryegrass and PAH-degrading bacteria. J. Hazard. Mater. 2011, 186, 1206–1217. [Google Scholar] [CrossRef]
- Liu, W.W.; Yin, R.; Lin, X.G.; Zhang, J.; Chen, X.M.; Li, X.Z.; Yang, T. Interaction of biosurfactant-microorganism to enhance phytoremediation of aged polycyclic aromatic hydrocarbons (PAHS) contaminated soils with alfalfa (Medicago sativa L.). Huan Jing Ke Xue 2010, 31, 1079–1084. [Google Scholar]
- Brinch, U.C.; Ekelund, F.; Jacobsen, C.S. Method for Spiking Soil Samples with Organic Compounds these include: Method for Spiking Soil Samples with Organic Compounds. Appl. Environ. Microbiol. 2002, 68, 1808–1816. [Google Scholar] [CrossRef]
- Mustafa, G.; Randoux, B.; Tisserant, B.; Fontaine, J.; Magnin-Robert, M.; Lounès-Hadj Sahraoui, A. Phosphorus supply, arbuscular mycorrhizal fungal species, and plant genotype impact on the protective efficacy of mycorrhizal inoculation against wheat powdery mildew. Mycorrhiza 2016, 26, 685–697. [Google Scholar] [CrossRef]
- McGonigle, T.P.; Miller, M.H.; Evans, D.G.; Fairchild, G.L.; Swan, J.A. A new method which gives an objective measure of colonization of roots by vesicular- arbuscular mycorrhizal fungi. New Phytol. 1990, 115, 495–501. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Letousey, P.; Spagnolo, A.; Rabenoelina, F.; Jacquens, L.; Mercier, L.; Clément, C.; Fontaine, F. Leaf stripe form of esca induces alteration of photosynthesis and defence reactions in presymptomatic leaves. Funct. Plant Biol. 2011, 38, 856–866. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: New York, NY, USA, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Meglouli, H.; Lounès-Hadj Sahraoui, A.; Magnin-Robert, M.; Tisserant, B.; Hijri, M.; Fontaine, J. Arbuscular mycorrhizal inoculum sources influence bacterial, archaeal, and fungal communities’ structures of historically dioxin/furan-contaminated soil but not the pollutant dissipation rate. Mycorrhiza 2018, 28, 635–650. [Google Scholar] [CrossRef]
- Dyke, P.H. Releases of Polychlorinated Dibenzo-p-Dioxins and Polychlorinated Dibenzofurans to Land and Water and with Products. In Persistent Organic Pollutants; The Handbook of Environmental Chemistry (Vol. 3 Series: Anthropogenic Compounds); Fiedler, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 30. [Google Scholar]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis, 2nd ed.; Chemical and Microbiological Properties; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 903–947. [Google Scholar]
- Adam, G.; Duncan, H. Development of a sensitive and rapid method for the measurement of total microbial activity using fluorescein diacetate (FDA) in a range of soils. Soil Biol. Biochem. 2001, 33, 943–951. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 2009, 10, e74014. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2013, 75, 7537–7541. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Knight, R. QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 2010, 5, 335–336. [Google Scholar] [CrossRef]
- Wu, F.Y.; Yu, X.Z.; Wu, S.C.; Lin, X.G.; Wong, M.H. Phenanthrene and pyrene uptake by arbuscular mycorrhizal maize and their dissipation in soil. J. Hazard. Mater. 2011, 187, 341–347. [Google Scholar] [CrossRef]
- Nwoko, C.O. Effect of arbuscular mycorrhizal AM fungi on the physiological performance of Phaseolus vulgaris grown under crude oil contaminated soil. J. Geosci. Environ. Prot. 2014, 2, 9–14. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, M. Effect of rhamnolipids on the uptake of PAHs by ryegrass. Environ. Pollut. 2008, 156, 46–52. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Klimova, S.Y.; Shestakov, A.I.; Botina, S.G.; Netrusov, A.I. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch. Microbiol. 2007, 188, 655–664. [Google Scholar] [CrossRef]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Boil. 2010, 2. [Google Scholar] [CrossRef]
- Urbaniak, M.; Zieliński, M.; Wyrwicka, A. The influence of the Cucurbitaceae on mitigating the phytotoxiccity and PCDD/PCDF content of soil amended with sewage sludge. Int. J. Phytoremediat. 2017, 19, 207–213. [Google Scholar] [CrossRef]
- Providenti, M.A.; Lee, H.; Trevors, J.T. Selected factors limiting the microbial degradation of recalcitrant compounds. J. Ind. Microbiol. 1993, 12, 379–395. [Google Scholar] [CrossRef]
- Grant, S.; Mortimer, M.; Stevenson, G.; Malcolm, D.; Gaus, C. Facilitated transport of dioxins in soil following unintentional release of pesticide-surfactant formulations. Environ. Sci. Technol. 2011, 45, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Chrzanowski, L.; Ławniczak, L.; Czaczyk, K. Why do microorganisms produce rhamnolipids? World J. Microbiol. Biotechnol. 2012, 28, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yin, R.; Lin, X.; Liu, W.; Chen, R.; Li, X. Interactive effect of biosurfactant and microorganism to enhance phytoremediation for removal of aged polycyclic aromatic hydrocarbons from contaminated soils. J. Health Sci. 2010, 56, 257–266. [Google Scholar] [CrossRef]
- Cheema, S.A.; Khan, M.I.; Tang, X.; Shen, C.; Farooq, M.; Chen, Y. Surfactant enhanced pyrene degradation in the rhizosphere of tall fescue Festuca arundinacea. Environ. Sci. Pollut. Res. 2016, 23, 18129–18136. [Google Scholar] [CrossRef]
- Manickam, N.; Bajaj, A.; Saini, H.S.; Shanker, R. Surfactant mediated enhanced biodegradation of hexachlorocyclohexane HCH isomers by Sphingomonas sp. NM05. Biodegradation 2012, 23, 673–682. [Google Scholar] [CrossRef]
- Camprubi, A.; Calvet, C.; Estaun, V. Growth enhancement of Citrus reshni after inoculation with Glomus intraradices and Trichoderma aureoviride and associated effects on microbial populations and enzyme activity in potting mixes. Plant Soil 1995, 173, 233–238. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, J.; Xiang, X. Impact of four plant species and arbuscular mycorrhizal AM fungi on polycyclic aromatic hydrocarbon PAH dissipation in spiked soil. Pol. J. Environ. Stud. 2013, 22, 1239–1245. [Google Scholar]
- Halden, R.U.; Halden, B.G.; Dwyer, D.F. Removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soils inoculated with Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 1999, 65, 2246–2249. [Google Scholar]
- Nam, I.H.; Kim, Y.M.; Schmidt, S.; Chang, Y.S. Biotransformation of 1,2,3-tri- and 1,2,3,4,7,8-hexachlorodibenzo-p- dioxin by Sphingomonas wittichii Strain RW1. Appl. Environ. Microbiol. 2006, 72, 112–116. [Google Scholar] [CrossRef]
- Wittich, R.M.; Wilkes, H.; Sinnwell, V.; Francke, W.; Fortnagel, P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 1992, 58, 1005–1010. [Google Scholar] [PubMed]
- Keim, T.; Francke, W.; Schmidt, S.; Fortnagel, P. Catabolism of 2,7-dichloro- and 2,4,8-trichlorodibenzofuran by Sphingomonas sp. strain RW1. J. Ind. Microbiol. Biotechnol. 1999, 23, 3359–3363. [Google Scholar] [CrossRef]
- Macek, T.; Macková, M.; Káš, J. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol. Adv. 2000, 18, 23–34. [Google Scholar] [CrossRef]
- Kuiper, I.; Lagendijk, E.L.; Bloemberg, G.V.; Lugtenberg, B.J.J. Rhizoremediation: A beneficial plant-microbe interaction. Mol. Plant Microbe. Interact. 2004, 17, 6–15. [Google Scholar] [CrossRef]
- Chaudhry, Q.; Blom-Zandstra, M.; Gupta, S.K.; Joner, E. Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. Res. 2005, 12, 34–48. [Google Scholar] [CrossRef]
- Yateem, A.; Al-Sharrah, T.; Bin-Haji, A. Investigation of microbes in the rhizosphere of selected grasses for rhizoremediation of hydrocarbon contaminated soils. Soil Sediment Contam. 2007, 16, 269–280. [Google Scholar] [CrossRef]
- Field, J.A.; Sierra-Alvarez, R. Microbial degradation of chlorinated dioxins. Chemosphere 2008, 71, 1005–1018. [Google Scholar] [CrossRef]
- Marco-Urrea, E.; Garcia-Romera, I.; Aranda, E. Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol. 2015, 32, 620–628. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Ni, Y.; Zhang, Q.; Zhao, L. Uptake by roots and translocation to shoots of polychlorinated dibenzo-p-dioxins and dibenzofurans in typical crop plants. Chemosphere 2009, 76, 740–746. [Google Scholar] [CrossRef]
- Tu, C.; Teng, Y.; Luo, Y.; Sun, X.; Deng, S.; Li, Z.; Liu, W.; Xu, Z. PCB removal, soil enzyme activities, and microbial community structures during the phytoremediation by alfalfa in field soils. J. Soils Sediments 2011, 11, 649–656. [Google Scholar] [CrossRef]
- Kaiya, S.; Rubaba, O.; Yoshida, N.; Yamada, T.; Hiraishi, A. Characterization of Rhizobium naphthalenivorans sp. nov. with special emphasis on aromatic compound degradation and multilocus sequence analysis of housekeeping genes. J. Gen. Appl. Microbiol. 2012, 58, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A. Biodiversity of dehalorespiring bacteria with special emphasis on polychlorinated biphenyl/dioxin dechlorinators. Microbes Environ. 2008, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Futamata, H.; Uchida, T.; Yoshida, N.; Yonemitsu, Y.; Hiraishi, A. Distribution of dibenzofuran-degrading bacteria in soils polluted with different levels of polychlorinated dioxins. Microbes Environ. 2004, 19, 172–177. [Google Scholar] [CrossRef]
- Huang, W.Y.; Ngo, H.H.; Lin, C.; Vu, C.T.; Kaewlaoyoong, A.; Boonsong, T.; Tran, H.T.; Bui, X.T.; Vo, T.D.H.; Chen, J.R. Aerobic co-composting degradation of highly PCDD/F-contaminated field soil. A study of bacterial community. Sci. Total Environ. 2019, 660, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S. Recent developments in microbial biotransformation and biodegradation of dioxins. J. Mol. Microbiol. Biotechnol. 2008, 15, 152–171. [Google Scholar] [CrossRef] [PubMed]
- Yuki, J.; Suzuki, M.; Miyamoto, H. Mold Capable of Degrading Dioxin, Degradation of Dioxin with the Use of the Same, Method for Producing Composts Capable of Degrading Dioxin and Method for Growing Plants. U.S. Patent 6,506,956 B1, 14 January 2003. [Google Scholar]
- Hammer, E.; Schauer, F. Fungal hydroxylation of dibenzofuran. Mycol. Res. 1997, 101, 433–436. [Google Scholar] [CrossRef]

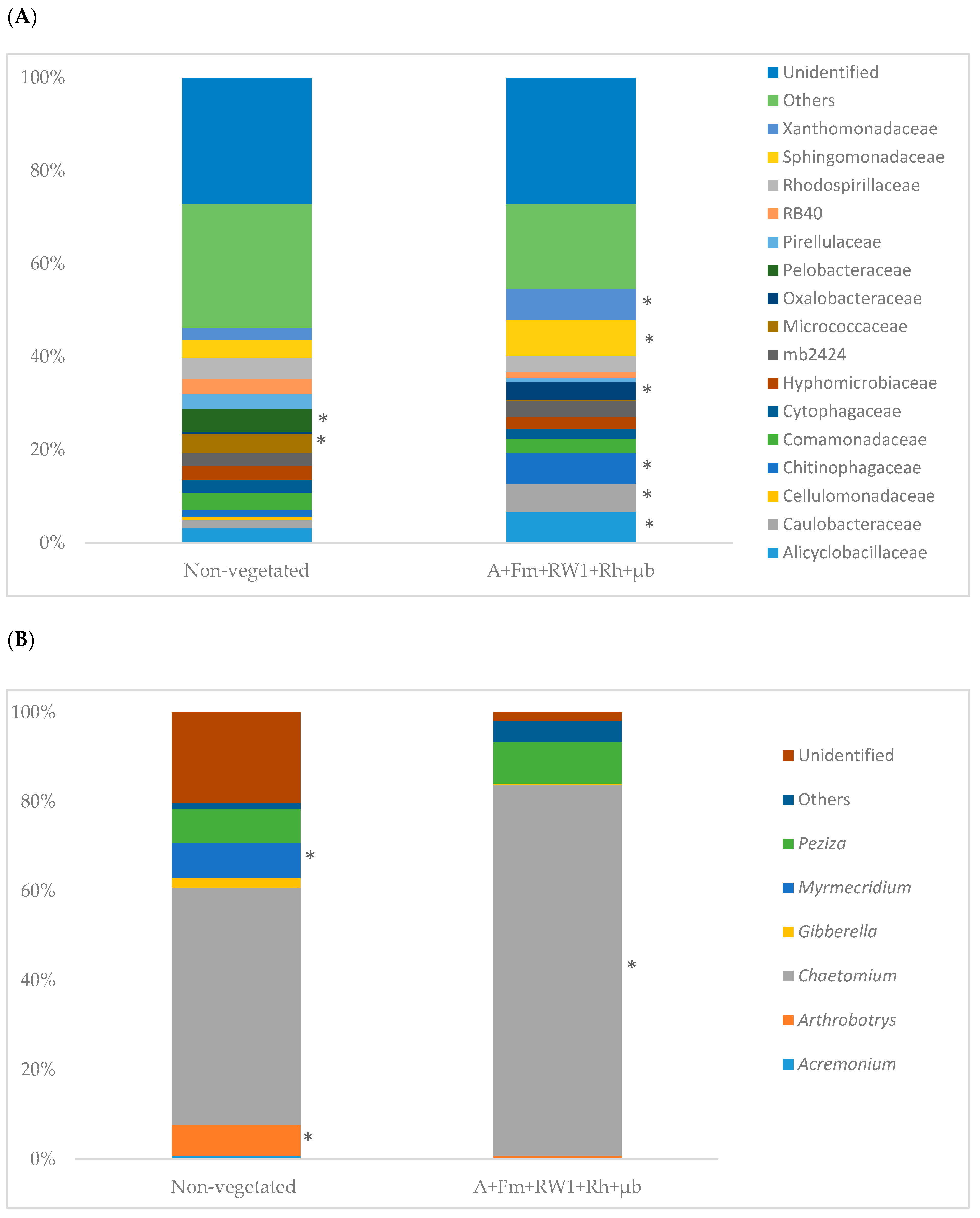
| Parameters | Values |
|---|---|
| Clay (g/kg) | 135 ± 10 |
| Silt: Fine particle (g/kg) | 149 ± 9 |
| Silt: Coarse particle (g/kg) | 431 ± 13 |
| Sand: Fine particle (g/kg) | 257 ± 16 |
| Sand: Coarse particle (g/kg) | 28 ± 6 |
| Organic C (g/kg) | 25.1 ± 6 |
| Total N (g/kg) | 1.53 ± 0.5 |
| Organic matter (g/kg) | 43.4 ± 8 |
| Total CaCO3 (g/kg) | <1 |
| P2O5 (g/kg) | 0.027 ± 0.027 |
| Mn (cmol+/kg) | 0.135 ± 0.008 |
| Ca (g/100g) | 0.565 ± 0.056 |
| Na (g/100g) | 0.697 ± 0.098 |
| Al (cmol+/kg) | 0.0469 ± 0.0293 |
| Fe (cmol+/kg) | 0.0089 ± 0.0013 |
| Si (g/100g) | 0.207 ± 0.065 |
| Mg (cmol+/kg) | 1.06 ± 0.3 |
| pH | 6.34 ± 0.035 |
| PCDD/F (ng/kg) | 208 ± 40 |
| Conditions | Control | Alfalfa (A) | F. mosseae (Fm) | S. wittichii (RW1) | Rhamnolipids (Rh) | Native Microbiota (µb) |
|---|---|---|---|---|---|---|
| Non-vegetated | x | |||||
| A | x | |||||
| A+Fm | x | x | ||||
| A+Fm+RW1 | x | x | x | |||
| A+Fm+RW1+Rh | x | x | x | x | ||
| A+Fm+RW1+Rh+µb | x | x | x | x | x |
| A | A+Fm | A+Fm+RW1 | A+Fm+RW1+Rh | A+Fm+RW1+Rh+µb | ||
|---|---|---|---|---|---|---|
| Mycorrhizal rate (%) | Total | 0 | 47 ± 2 a | 39 ± 4 ab | 30 ± 8 b | 40 ± 6 ab |
| Arbuscular | 0 | 12 ± 2 b | 7 ± 1 b | 7 ± 3 ab | 10 ± 1 b | |
| Vesicular | 0 | 31 ± 3 a | 25 ± 8 ab | 20 ± 6 b | 23 ± 2 ab | |
| Plant dry weight (mg plant−1) | Shoot dry weight | 97 ± 1 a | 279 ± 1.6 d | 171 ± 30 b | 191 ± 15 b | 207 ± 18 bc |
| Root dry weight | 60 ± 12 ab | 67 ± 19 b | 76 ± 3 bc | 39 ± 8 a | 55 ± 2 ab |
| Cells | Enzymes | A+Fm+RW1+Rh+µb (%) |
|---|---|---|
| Lung Cells (Beas2B) | LDH activity vs. control | −44 ± 24 (p < 0.0001) |
| MDH activity vs. control | +5 ± 1 (p = 0.751) | |
| Liver Cells (HepG2) | LDH activity vs. control | −5 ± 1 (p = 0.0001) |
| MDH activity vs. control | +18 ± 9 (p < 0.0001) |
| Conditions | Dehydrogenase (µg g−1 of Soil/24 h) | Fluorescein Diacetate Hydrolase (µg g−1 of Soil) |
|---|---|---|
| Non-vegetated | 65 ± 3 a | 5.0 ± 0.1 a |
| A | 196 ± 28 bcd | 17.0 ± 6.0 b |
| A+Fm | 288 ± 94 cd | 51.0 ± 10.0 c |
| A+Fm+RW1 | 186 ± 36 bc | 55.0 ± 7.0 c |
| A+Fm+RW1+Rh | 173 ± 22 b | 25.0 ± 4.0 b |
| A+Fm+RW1+Rh+µb | 281 ± 31 d | 70.0 ± 7.0 d |
| Observed OTUs | Chao1 Richness Estimate | Shannon Diversity Index | ||
|---|---|---|---|---|
| Bacteria | Non-vegetated | 481 ± 23 a | 488 ± 34 a | 7.10 ± 0.14 a |
| A+Fm+RW1+Rh+µb | 618 ± 57 b | 657 ± 51b | 7.50 ± 0.13 b | |
| Archaea | Non-vegetated | 9 ± 1 a | 8 ± 1a | 0.90 ± 0.42 a |
| A+Fm+RW1+Rh+µb | 7 ± 2 a | 7 ± 1 a | 1.30 ± 0.05 a | |
| Fungi | Non-vegetated | 604 ± 180 a | 3890 ± 406 a | 1.21 ± 0.80 a |
| A+Fm+RW1+Rh+µb | 644 ± 84 a | 4108 ± 378 a | 2.44± 0.26 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meglouli, H.; Fontaine, J.; Verdin, A.; Magnin-Robert, M.; Tisserant, B.; Hijri, M.; Lounès-Hadj Sahraoui, A. Aided Phytoremediation to Clean Up Dioxins/Furans-Aged Contaminated Soil: Correlation between Microbial Communities and Pollutant Dissipation. Microorganisms 2019, 7, 523. https://doi.org/10.3390/microorganisms7110523
Meglouli H, Fontaine J, Verdin A, Magnin-Robert M, Tisserant B, Hijri M, Lounès-Hadj Sahraoui A. Aided Phytoremediation to Clean Up Dioxins/Furans-Aged Contaminated Soil: Correlation between Microbial Communities and Pollutant Dissipation. Microorganisms. 2019; 7(11):523. https://doi.org/10.3390/microorganisms7110523
Chicago/Turabian StyleMeglouli, Hacène, Joël Fontaine, Anthony Verdin, Maryline Magnin-Robert, Benoît Tisserant, Mohamed Hijri, and Anissa Lounès-Hadj Sahraoui. 2019. "Aided Phytoremediation to Clean Up Dioxins/Furans-Aged Contaminated Soil: Correlation between Microbial Communities and Pollutant Dissipation" Microorganisms 7, no. 11: 523. https://doi.org/10.3390/microorganisms7110523
APA StyleMeglouli, H., Fontaine, J., Verdin, A., Magnin-Robert, M., Tisserant, B., Hijri, M., & Lounès-Hadj Sahraoui, A. (2019). Aided Phytoremediation to Clean Up Dioxins/Furans-Aged Contaminated Soil: Correlation between Microbial Communities and Pollutant Dissipation. Microorganisms, 7(11), 523. https://doi.org/10.3390/microorganisms7110523







