Microbiological Assessment of the Quality of Some Commercial Products Marketed as Lactobacillus crispatus-Containing Probiotic Dietary Supplements
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Viable Cell Number Counts
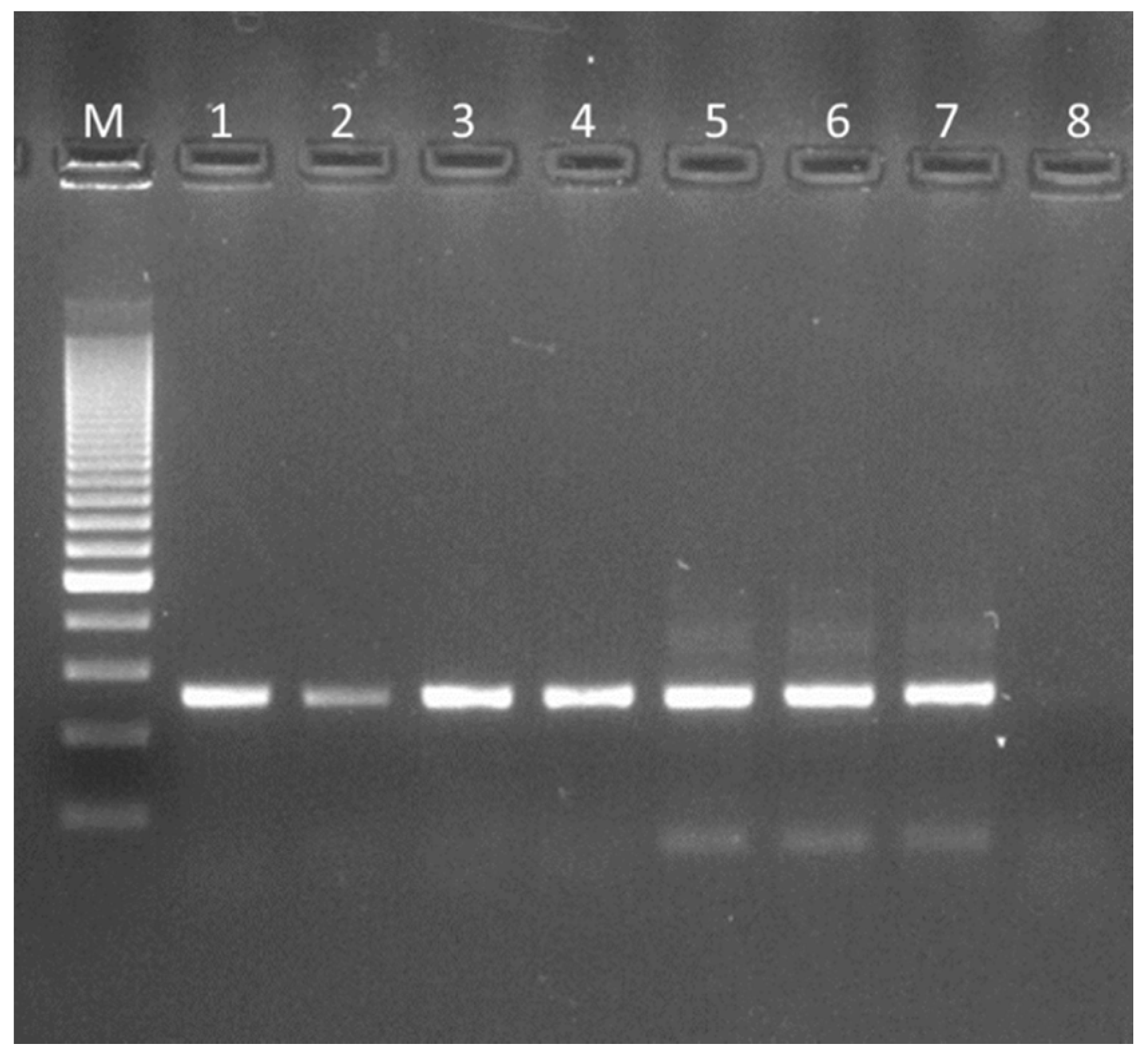
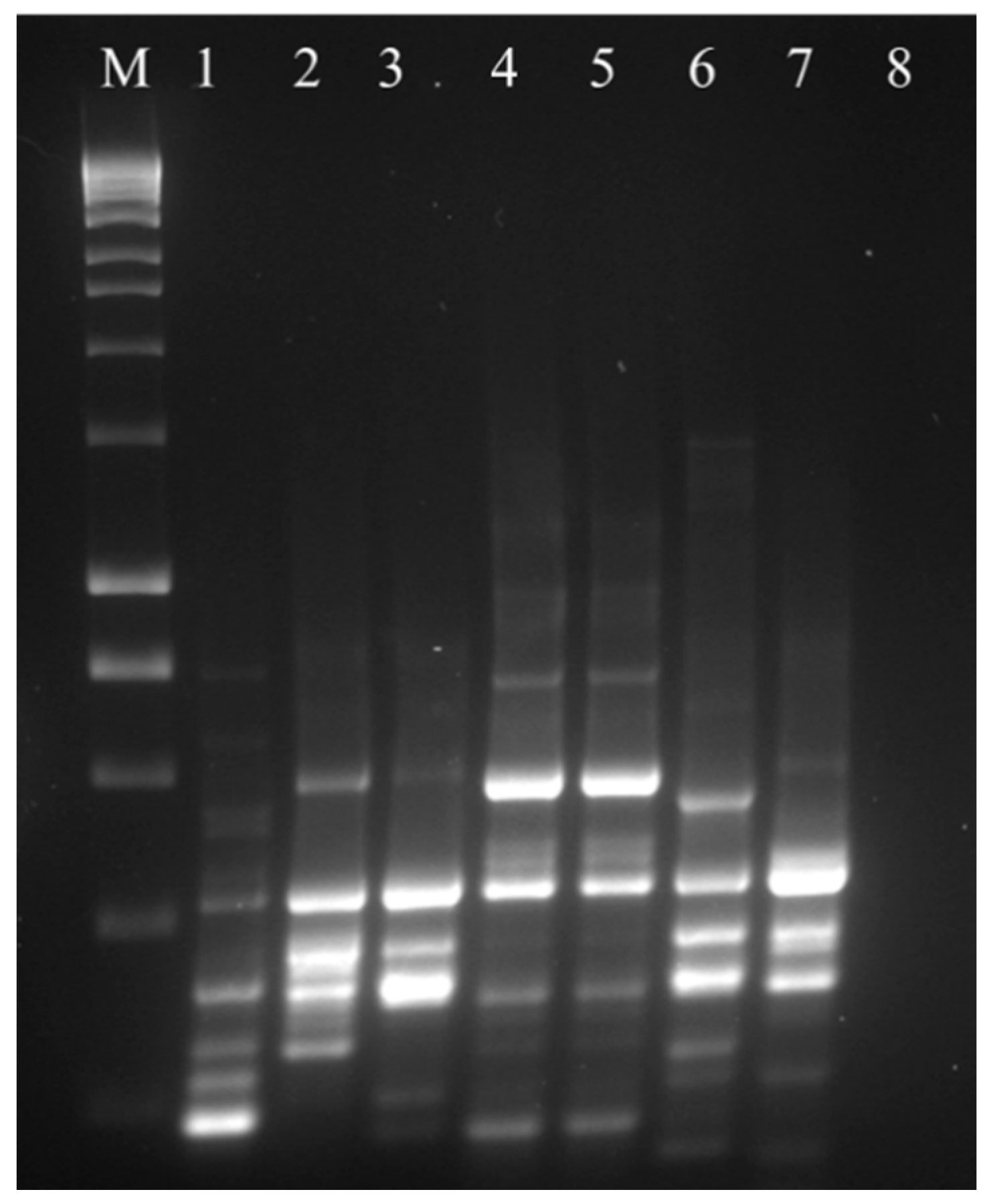
2.3. Isolation and Extraction of L. crispatus Colony DNA, 16S rRNA Gene Amplification and Molecular Typing
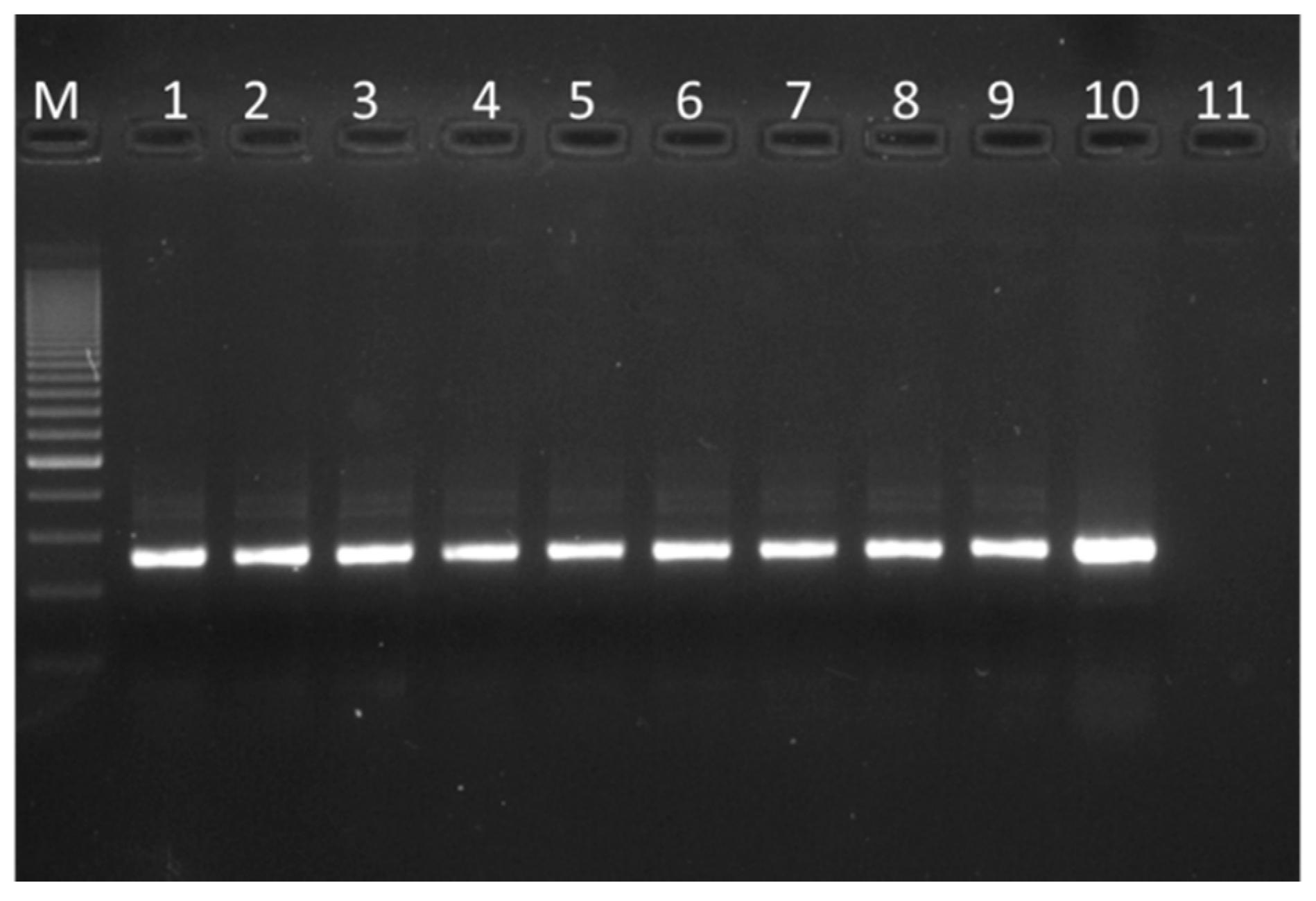
2.4. Total DNA Extraction from Finished Products
3. Results
3.1. Viable Cell Number Counts
3.2. 16 S rRNA Gene Amplification and Molecular Typing of Viable Colonies
3.3. 16 S rRNA Gene Amplification of the Finished Products
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sanders, M.E.; Merenstein, D.; Merrifield, C.A.; Hutkins, R. Probiotics for human use. Nutr. Bull. 2018, 43, 212–225. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Kolaček, S.; Hojsak, I.; Berni Canani, R.; Guarino, A.; Indrio, F.; Orel, R.; Pot, B.; Shamir, R.; Szajewska, H.; Vandenplas, Y.; et al. Commercial Probiotic Products: A Call for Improved Quality Control. A Position Paper by the ESPGHAN Working Group for Probiotics and Prebiotics. J. Pediatr. Gastroenterol. Nutr. 2017, 65, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Rodighiero, V.; Celeste, T.; Rovetto, L.; De Vecchi, E. Microbiological evaluation of commercial probiotic products available in the USA in 2009. J. Chemother. 2010, 22, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Martin, H. Assessment of commercial probiotic bacterial contents and label accuracy. Can. Vet. J. 2011, 52, 43–46. [Google Scholar]
- Morovic, W.; Hibberd, A.A.; Zabel, B.; Barrangou, R.; Stahl, B. Genotyping by PCR and high-throughput sequencing of commercial probiotic products reveals composition biases. Front. Microbiol. 2016, 7, 1747. [Google Scholar] [CrossRef]
- Aureli, P.; Fiore, A.; Scalfaro, C.; Casale, M.; Franciosa, G. National survey outcomes on commercial probiotic food supplements in Italy. Int. J. Food Microbiol. 2010, 137, 265–273. [Google Scholar] [CrossRef]
- Celandroni, F.; Vecchione, A.; Cara, A.; Mazzantini, D.; Lupetti, A.; Ghelardi, E. Identification of Bacillus species: Implication on the quality of probiotic formulations. PLoS ONE 2019, 14, e0217021. [Google Scholar] [CrossRef]
- Patrone, V.; Molinari, P.; Morelli, L. Microbiological and molecular characterization of commercially available probiotics containing Bacillus clausii from India and Pakistan. Int. J. Food Microbiol. 2016, 237, 92–97. [Google Scholar] [CrossRef]
- Jackson, S.A.; Schoeni, J.L.; Vegge, C.; Pane, M.; Stahl, B.; Bradley, M.; Goldman, V.S.; Burguière, P.; Atwater, J.B.; Sanders, M.E. Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 2019, 10, 739. [Google Scholar] [CrossRef]
- Schoeni, J.L. Probiotics. In Compendium of Methods for the Microbiological Examination of Foods, 5th ed.; Salfinger, Y., Tortorello, M.L., Eds.; American Public Health Association: Washington, DC, USA, 2015; pp. 232–286. [Google Scholar]
- Hansen, S.J.Z.; Morovic, W.; DeMeules, M.; Stahl, B.; Sindelar, C.W. Absolute enumeration of probiotic strains Lactobacillus acidophilus NCFM® and Bifidobacterium animalis subsp. lactis Bl-04® via chip-based digital PCR. Front. Microbiol. 2018, 9, 704. [Google Scholar] [PubMed]
- Lugli, G.A.; Mangifesta, M.; Mancabelli, L.; Milani, C.; Turroni, F.; Viappiani, A.; van Sinderen, D.; Ventura, M. Compositional assessment of bacterial communities in probiotic supplements by means of metagenomic techniques. Int. J. Food Microbiol. 2019, 294, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef] [PubMed]
- Lepargneur, J.P. Lactobacillus crispatus as biomarker of the healthy vaginal tract. Ann. Biol. Clin. 2016, 74, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Hütt, P.; Lapp, E.; Štšepetova, J.; Smidt, I.; Taelma, H.; Borovkova, N.; Oopkaup, H.; Ahelik, A.; Rööp, T.; Hoidmets, D.; et al. Characterisation of probiotic properties in human vaginal lactobacilli strains. Microb. Ecol. Health. Dis. 2016, 27, 30484. [Google Scholar] [CrossRef]
- Toscano, M.; De Grandi, R.; Bortolin, M.; Drago, L. Biofilm production and adhesion to intestinal cells: Characterization of six lactobacillus strains isolated from Italian probiotic products. Int. J. Probiotics 2017, 12, 153–158. [Google Scholar]
- Sierra-Arguello, Y.M.; Faulkner, O.; Tellez, G.; Hargis, B.M.; Pinheiro do Nascimento, V. The use of FTA cards for transport and detection of gyrA mutation of Campylobacter jejuni from poultry. Poult. Sci. 2016, 95, 798–801. [Google Scholar] [CrossRef]
- Song, Y.; Kato, N.; Liu, C.; Matsumiya, Y.; Kato, H.; Watanabe, K. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 2000, 187, 167–173. [Google Scholar] [CrossRef]
- Cocconcelli, P.S.; Parisi, M.G.; Senini, L.; Bottazzi, V. Use of RAPD and 16S rDNA sequencing for the study of Lactobacillus population dynamics in natural whey culture. Lett. Appl. Microbiol. 1997, 25, 8–12. [Google Scholar] [CrossRef]
- Fontana, C.; Cocconcelli, P.S.; Vignolo, G. Monitoring the bacterial population dynamics during fermentation of artisanal Argentinean sausages. Int. J. Food Microbiol. 2005, 103, 131–142. [Google Scholar] [CrossRef]
- Payne, A.N.; Chassard, C.; Zimmermann, M.; Müller, P.; Stinca, S.; Lacroix, C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr. Diabetes 2011, 1, e12. [Google Scholar] [CrossRef] [PubMed]
- Verdenelli, M.C.; Coman, M.M.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Cresci, A. Evaluation of antipathogenic activity and adherence properties of human Lactobacillus strains for vaginal formulations. J. Appl. Microbiol. 2014, 116, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Strus, M.; Chmielarczyk, A.; Kochan, P.; Adamski, P.; Chełmicki, Z.; Chełmicki, A.; Pałucha, A.; Heczko, P.B. Studies on the effects of probiotic Lactobacillus mixture given orally on vaginal and rectal colonization and on parameters of vaginal health in women with intermediate vaginal flora. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 163, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Reid, G. The development of probiotics for women's health. Can. J. Microbiol. 2017, 63, 269–277. [Google Scholar] [CrossRef]
- L 117. Off. J. Eur. Union 2017, 60, 1–175.
- Voltan, S.; Martines, D.; Elli, M.; Brun, P.; Longo, S.; Porzionato, A.; Macchi, V.; D'Incà, R.; Scarpa, M.; Palù, G.; et al. Lactobacillus crispatus M247-derived H2O2 acts as a signal transducing molecule activating peroxisome proliferator activated receptor-gamma in the intestinal mucosa. Gastroenterology 2008, 135, 1216–1227. [Google Scholar] [CrossRef]
- Siciliano, R.A.; Cacace, G.; Mazzeo, M.F.; Morelli, L.; Elli, M.; Rossi, M.; Malorni, A. Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim. Biophys. Acta 2008, 1784, 335–342. [Google Scholar]
- Voltan, S.; Castagliuolo, I.; Elli, M.; Longo, S.; Brun, P.; D'Incà, R.; Porzionato, A.; Macchi, V.; Palù, G.; Sturniolo, G.C.; et al. Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin. Vaccine Immunol. 2007, 14, 1138–1148. [Google Scholar] [CrossRef]
- Castagliuolo, I.; Galeazzi, F.; Ferrari, S.; Elli, M.; Brun, P.; Cavaggioni, A.; Tormen, D.; Sturniolo, G.C.; Morelli, L.; Palù, G. Beneficial effect of auto-aggregating Lactobacillus crispatus on experimentally induced colitis in mice. FEMS Immunol. Med. Microbiol. 2005, 43, 197–204. [Google Scholar] [CrossRef][Green Version]
- Marcotte, H.; Ferrari, S.; Cesena, C.; Hammarström, L.; Morelli, L.; Pozzi, G.; Oggioni, M.R. The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J. Appl. Microbiol. 2004, 97, 749–756. [Google Scholar] [CrossRef]
- Kirjavainen, P.V.; Ouwehand, A.C.; Isolauri, E.; Salminen, S.J. The ability of probiotic bacteria to bind to human intestinal mucus. FEMS Microbiol. Lett. 1998, 167, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Cesena, C.; Morelli, L.; Alander, M.; Siljander, T.; Tuomola, E.; Salminen, S.; Mattila-Sandholm, T.; Vilpponen-Salmela, T.; von Wright, A. Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. J. Dairy Sci. 2001, 84, 1001–1010. [Google Scholar] [CrossRef]
- Di Pierro, F.; Bertuccioli, A.; Cattivelli, D.; Soldi, S.; Elli, M. Lactobacillus crispatus M247: A possible tool to counteract CST IV. Nutrafoods 2018, 17, 169–172. [Google Scholar]
| Product | Pharmaceutical Form | L. crispatus Strain | Other Bacterial Species | Market | Total Dose Declared (CFU/gr) | L. crispatus Dose Declared (CFU/gr) |
|---|---|---|---|---|---|---|
| (A) | Stick | M247 | / | Italy/NS | not less than 2 × 1010 | not less than 2 × 1010 |
| (B) | Sachet | LBV10 | L. gasseri LBV162 | Italy/NS | not less than 2 × 109 | not less than 1 × 109 |
| (C) | IMC6075 | L. rhamnosus IMC501, | Italy/NS | 4 × 1011 | 1 × 1011 | |
| Capsule | L. jensenii IMC813S, | |||||
| L. reuteri LR92 | ||||||
| (D) | Capsule | IP174178 | / | France/NS | 2.8 × 108 | 2.8 × 108 |
| (E) | IP174178 | / | France/MD | 4 × 108 | 4 × 108 | |
| (F) | KS127 | L.rhamosus LB21, | Italy/NS | 2 × 1010 | 2 × 109 | |
| Capsule | L. plantarum LB931, | |||||
| B. lactis Bi1 | ||||||
| (G) | IMC607S | L. gasseri LG050 | Italy/NS | 5.4 × 109 | 2.7 × 109 | |
| (H) | Sachet | P17631 | L. rhamnosus ATCC53103 | Italy/NS | ND | ND |
| (I) | Not indicated | L. rhamnosus, | Italy/MD | 4 × 1011 | 1 × 1011 | |
| Capsule | L. jensenii, | |||||
| L. reuteri |
| Log 10(CFU/gr) | ||||
|---|---|---|---|---|
| Product | L. crispatus Strain | MRS | MRS+Clinda/Ciproflox | MRS+Vancomycin |
| (A) | M247 | 10.4 ± 0.2 | 10.4 ± 0.4 | ND |
| (B) | LBV10 | 9.4 ± 0.1 | 8.9 ± 0.1 | ND |
| (C) | IMC6075 | 8.0 ± 0.1 | NG | 7.5 ± 0.4 |
| (D) | IP174178 | 8.1 ± 0.3 | 8.1 ± 0.3 | ND |
| (E) | IP174178 | 8.0 ± 0.2 | 8.0 ± 0.3 | ND |
| (F) | KS127 | 9.9 ± 0.1 | 8.0 ± 0.3 | 9.7 ± 0.3 |
| (G) | IMC607S | 2.0 ± 0.1 | NG | NG |
| (H) | P17631 | 6.7 ± 0.2 | NG | 6.7 ± 0.5 |
| (I) | NI | 11 ± 0.1 | NG | 10 ± 0.4 |
| Product | L. Crispatus Strain | Weight (g) | Daily Dose | Total probiotics-Measured Viability Per Daily Dose | Total probiotics-Declared Daily Dose | Potential L. crispatus Per Daily Dose | Declared L. crispatus Per Daily Dose |
|---|---|---|---|---|---|---|---|
| (A) | M247 | 1 g/stick | 1 stick | 10.4 | 10 | 10.41 | 10 |
| (B) | LBV10 | 2.8 g/sachet | 1 sachet | 9.7 | 9.3 | 9.22 | 9 |
| (C) | IMC6075 | 0.35 g/capsule | 1 capsule | 7.5 | 9.6 | NG | 9 |
| (D) | IP174178 | 0.45g/capsule | 1 capsule | 7.8 | 8 | 7.83 | 8 |
| (E) | IP174178 | 0.35 g/capsule | 1–2 capsule | 7.5 | 9 | 7.54 | 9 |
| (F) | KS127 | 0.5 g/capsule | 1 capsule | 9.6 | 9.8 | 7.75 | 9 |
| (G) | IMC607S | 0.375 g/capsule | 1 capsule | 1.5 | 9.3 | NG | 9 |
| (H) | P17631 | 1.5 g/sachet | 1 sachet | 6.3 | NL | NG | NL |
| (I) | NI | 0.35 g/capsule | 1 capsule | 10.6 | 10.4 | NG | 9.7 |
| Product | L. crispatus Strain | N |
|---|---|---|
| A | M247 | 7 |
| B | LBV10 | 0 |
| C/G | IMC607S | 0 |
| D/E | IP174178 | 0 |
| F | KS127 | 0 |
| H | P17631 | 0 |
| I | Type of strain not indicated | N. F.* |
| X | CTV-05 | 10 ^ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pierro, F.; Polzonetti, V.; Patrone, V.; Morelli, L. Microbiological Assessment of the Quality of Some Commercial Products Marketed as Lactobacillus crispatus-Containing Probiotic Dietary Supplements. Microorganisms 2019, 7, 524. https://doi.org/10.3390/microorganisms7110524
Di Pierro F, Polzonetti V, Patrone V, Morelli L. Microbiological Assessment of the Quality of Some Commercial Products Marketed as Lactobacillus crispatus-Containing Probiotic Dietary Supplements. Microorganisms. 2019; 7(11):524. https://doi.org/10.3390/microorganisms7110524
Chicago/Turabian StyleDi Pierro, Francesco, Valeria Polzonetti, Vania Patrone, and Lorenzo Morelli. 2019. "Microbiological Assessment of the Quality of Some Commercial Products Marketed as Lactobacillus crispatus-Containing Probiotic Dietary Supplements" Microorganisms 7, no. 11: 524. https://doi.org/10.3390/microorganisms7110524
APA StyleDi Pierro, F., Polzonetti, V., Patrone, V., & Morelli, L. (2019). Microbiological Assessment of the Quality of Some Commercial Products Marketed as Lactobacillus crispatus-Containing Probiotic Dietary Supplements. Microorganisms, 7(11), 524. https://doi.org/10.3390/microorganisms7110524








